【原创】也谈Real-time PCR引物设计
丁香园论坛
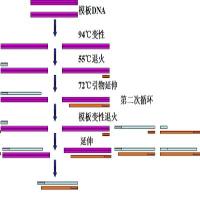
先谈引物设计原则,再讲实施方法。
引物设计不仅对引物有要求,也对PCR target有要求。
Design guidelines for primers:
Primers:
1 length
18-24
2 GC content
35%-65%(ideally 40-60%)
3 Tm
60-70°C (ideally 65°C), so that annealing temperature is 58-62°C
ΔTm (forward primer and reverse primer)<4°C
4 3’ end
Avoid 3’ end T (allow mismatching), G or C is better
No more than 3 G+C at the 3’ end in 5 nucleotides
Avoid runs of identical nucleotides, especially of 3 or more Gs or Cs at the 3’ end
5 Avoid hairpins
6 Avoid complementarity between the primers, especially at the 3’ ends of the primers (2 or more bases) to avoid primer-dimers
7 Take into account alternative splicing
8 Take into account into known SNPs
9 If Oligo-dT is used in RT, keep close to Poly-A tail
10 Design intron spanning or franking primers to avoid co-amplification of genomic DNA, which is only possible in multiple exon genes. (For single exon genes, perform DNase I treatment of RNA samples)
11 Use software (Oligo) to detect false priming sites in the template mRNA sequence to enhance the specificity of priming and avoid the low-complexity regions
Amplicon
1 SYBR green I assays:
80-200 bp (ideally 80-150). Alternative length ranges are often used to satisfied other requirement, but remember that shorter amplicons would give higher PCR efficiencies
2 GC content
30-80%(ideally 40-60%)
3 Avoid secondary structures in the amplicon (check with mfold)
4 Check if generate amplicon is unique
Discriminate cDNA from genomic DNA
Discriminate cDNA from pseudo-genes. If pseudo-genes exit, multi-align the sequences between the gene and it’s pseudo-genes to make sure the difference region to help you to design the primers