siRNA Design
互联网
siRNA Design
RNAi target selection rules :
Targeted regions on the cDNA sequence of a targeted gene should be located 50-100 nt downstream of the start codon (ATG).
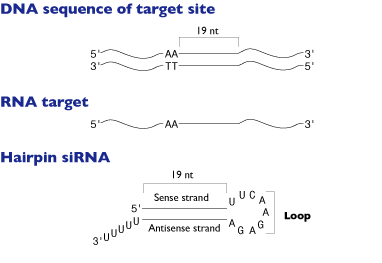
Search for sequence motif AA(N19 )TT or NA(N21 ), or NAR(N17 )YNN, where N is any nucleotide, R is purine (A, G) and Y is pyrimidine (C, U).
Avoid targeting introns, since RNAi only works in the cytoplasm and not within the nucleus.
Avoid sequences with > 50% G+C content.
Avoid stretches of 4 or more nucleotide repeats.
Avoid 5URT and 3UTR, although siRNAs targeting UTRs have successfully induced gene inhibition.
Avoid sequences that share a certain degree of homology with other related or unrelated genes.
How to obtain a cDNA sequence for target selection
Before finding a RNAi target on the gene of your interest, first you have to get its mRNA sequence or sequence accession number as some siRNA design tools can take accession number as input. It is recommended to use the gene's RefSeq from NCBI , since the RefSeq represents non-redundant, curated and validated sequences. RefSeq mRNA sequences have unique accession numbers which start with NM or XM, followed by 6 digits. For example, NM_123456 (curated mRNA sequence) or XM_0123456 (model mRNA sequence predicted by genome sequence analysis). There are several ways of querying RefSeq.
Search LocusLink by gene name or symbol at http://www.ncbi.nlm.nih.gov/LocusLink/ . Once the locus of your gene is found, scroll down to the "NCBI Reference Sequence (RefSeq)" section and look for mRNA.
Search Entrez Gene at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene , and select the right gene of desired organism. Once the page for the gene is shown, scroll down to the "NCBI Reference Sequence (RefSeq)" and look for mRNA.
Search Nucleotide database using Entrez query tool at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide and use Entrez Limits settings to restrict your query to the RefSeq database only
select "RefSeq" from the "Only from" menu, this restricts the query to the RefSeq collection
select "mRNA" from the "Molecule" menu, this restricts the query to mRNA RefSeq records
Homology search
The RNAi targeted region on the mRNA sequence of a gene should not share significant homology with other genes or sequences in the genome, therefore, homology search is essential to minimize off-target effects. Although most siRNA design tools provide BLAST option, some simply use NCBI BLAST tools which sometimes are quite slow. Here are some BLAST tools for homology search.
NCBI Blast tool: Nucleotide-nucleotide BLAST (blastn) or Search for short, nearly exact matches
Blat tool on UCSC Genome Website http://genome.ucsc.edu/cgi-bin/hgBlat
Ensembl Blast http://www.ensembl.org/Multi/blastview
Examples of RNAi target selection
References
1. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001 May 24;411(6836):494-8.
2. Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001 Jan 15;15(2):188-200.