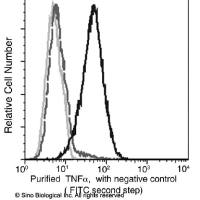
Measurement of TNF and iNOS mRNA Using cDNA-Equalized Reverse Transcriptase PCR
互联网
596
Since the polymerase chain reaction (PCR) for DNA amplification was first introduced in 1985 (1 ), the combination of reverse transcription with subsequent PCR amplification of the cDNA (RT-PCR) has been an increasingly utilized technique to analyze gene expression (2 ,3 ). In order for this procedure to be reasonably quantitative, however, appropriate controls must be applied to all steps, including the quantitation of the original RNA, the reverse transcription, and the PCR itself. Several investigators have published methods on quantitative RT-PCR that involve varying cDNA input into the PCR (3 –7 ), varying cycle number (3 ,4 ,8 ,9 ), or the use of a competitive template as an internal standard (10 ,11 ). However, only a few of the competitive PCR methods take into account the efficency of the reverse transcription phase of RT-PCR (4 ,7 ,11 ), which may vary from 10-50% (12 ,13 ). In the former methods, it would also be necessary to amplify another control gene in parallel (e.g., actin) to control for RNA input and reverse transcription.