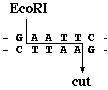Restriction Fragment Length Polymorphism (RFLP) Analyses: Distinguishing African and European Honey Bees
互联网
543
With the discovery of bacterial “restriction endonucleases” (1 ), it became possible to fractionate the long DNA molecule, so that specific regions could be analyzed. These enzymes cut DNA at specific short sequences, commonly consisting of four to six nucleotides (2 ). DNA fragments of discrete lengths are generated, which can be separated by electrophoresis through a gel matrix. In fragmenting DNA, restriction enzymes can be used as a reflection of the nucleotide sequences that they recognize, allowing genetic distinction based on a small percentage of the genome, without having to sequence the DNA (3 ). If a sequence recognized by a restriction enzyme is altered by a substitution of any of the nucleotides, it would not be cleaved. Likewise, nucleotide changes may create new sites. Such differences, resulting from genetic divergence, would be manifested in the length of fragments generated by the enzymes and separated by electrophoresis. Fragments generated by other enzymes would reflect positions of different restriction sites along the same region of DNA and would reveal changes in the additional sets of nucleotides. Changes in restriction fragment patterns result not only from point mutations but also from deletions, insertions, or duplications that alter the length of fragments (4 –6 ). In the nuclear genome, polymorphisms also reflect different locations of the same sequence (repetitive or transposable DNA), owing to different flanking DNA (7 ,8 ).