Inverse PCRI. Genomic DNA Prep• from 5 ml culture resuspend in 50 µl TEII. Digestions genomic DNA 5 µl 10x of appropriate NEB buffer 5 µl 0.5 µg/µl RNase 1 µl H2O 38 µl restriction enzyme 1 µl • Use ...
I. Quick Fly Genomic DNA Prep Standard fly mini prep (30 flies) resuspended in 150 ul TE 1) Collect 30 anesthetized flies in eppendorf tube and freeze at -80°. 2) Grind flies in 200 µl Buffer A with ...
Multiplex PCR is a variant of PCR which enabling simultaneous amplification of many targets of interest in one reaction by using more than one pair of primers. Since its first description in 1988 by C ...
PCR and multiplex PCR guide Table below lists the parameters influencing the PCR reaction and indicates some PCR and multiplex PCR applications. Clicking on an item in the table will take you to a det ...
Troubleshooting for PCR and multiplex PCRTroubleshooting discussion is based on the PCR protocol as described in the table below. All reactions are run for 30 cycles.COMPONENTVOLUMEFINAL CONCENTRATION ...
Microdeletion screeningOne application of multiplex PCR is microdeletion screening. This can be applied to the X and Y chromosomes (male genomic DNA) or to hybrid cell lines (rodent-human) containing ...
Standard multiplex mixturesOver 75 primer pairs were chosen and a number of multiplex mixtures were designed and used for different purposes. Examples of all multiplex mixes are presented below.(All u ...
End repair: Add 5-10 units of T4 DNApol and incubate at 37C for 5 minutes. Make solution to 2.5 M NH4OAc and add 1 volume of 95% ethanol; let sit for 5 minutes at RT and spin at 14K x g for 20 minutes ...
When designing PCR experiments in which the synthesized DNA fragment is to be subsequently digested with a RE it is very important to determine how many extra nucleotides should be added to the 5’-en ...
PURIFICATION OF PCR PRODUCTS WITH SEPHADEXPlace the sephadex measuring plate (MultiScreen ?Column Loader) on a clean piece of saran wrap. Pour some sephadex onto the plate (takes about 3.3 g). Scrape ...
Protocol for Real-Time RT-PCRThis protocol describes the detailed experimental procedure for real-time RT-PCR using SYBR Green I as mentioned in Xiaowei Wang and Brian Seed (2003) A PCR primer bank fo ...
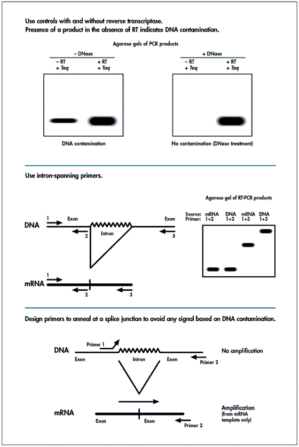
Detecting DNA Contamination in RT-PCR
Avoiding DNA Contamination in RT-PCR A frequent cause of concern among investigators performing quantitative RT-PCR is inaccurate data due to DNA contamination in RNA preparations. Although DNA contam ...
Designing PCR programsBasic Principles (see also Page 01) The requirement of an optimal PCR reaction is to amplify a specific locus without any unspecific by-products. Therefore annealing needs to tak ...
Single Primer ("Semi-Random") PCRJuly 26 2000 ECKDescriptionSingle primer PCR allows amplification from known to unknown regions in chromosomes phage plasmids large PCR products and other sources of D ...
This procedure was first described by Bertrand et al (Proceedings of the National Academy of Science USA (1993) Vol. 90 pp. 3496-3500 and Nucleic Acids Research (1994) Vol. 22 pp. 293-300) to demonstr ...
Protocol for competitive RT-PCR For quantifying mRNA we use a competitive RT-PCR protocol with internal standard RNAs. These are added in a defined quantity to the RNA sample prior to the RT reaction. ...
Successful RT-PCR requires a high quality intact RNA template. Use the following guidelines to help prepare this template: To minimize the activity of RNases that are released during cell lysis inclu ...
RT-PCR AnalysisSolutions10X RT Buffer10X PCR Buffer100 mM Tris pH 9.0500 mM KCl1% Triton X-10025 mM MgCl2use at a concentration of 1.5 mMLysis Solution4M GuSCN 250 g guanidine thiocyanate25mM Na citr ...
Question 1.What are the differences among RNase H RNase A RNase B and RNase C?2.In your cDNA kits RNase H is added in the second strand reaction to produce more nicked RNA as primers for DNA synthes ...