大家都在搜
手机验证

伯豪生物
4个空间转录组送2个单细胞(核)转录组+4个bulk RNA测序
以电话询价为准

风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
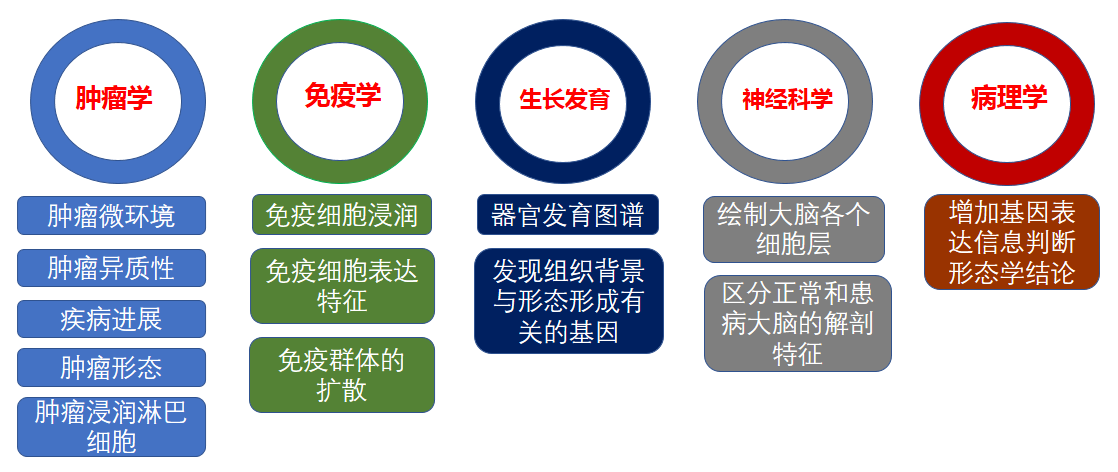
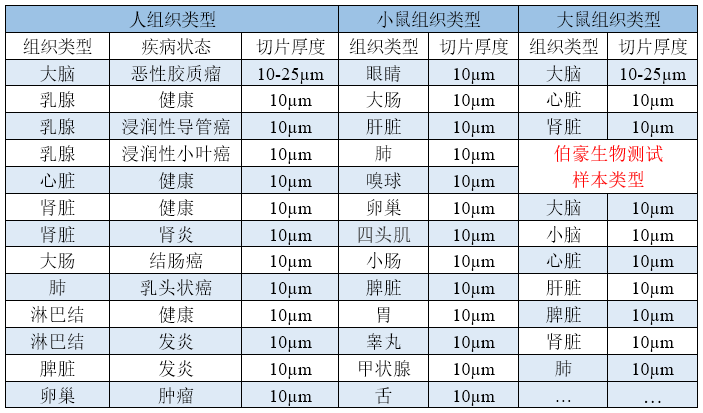
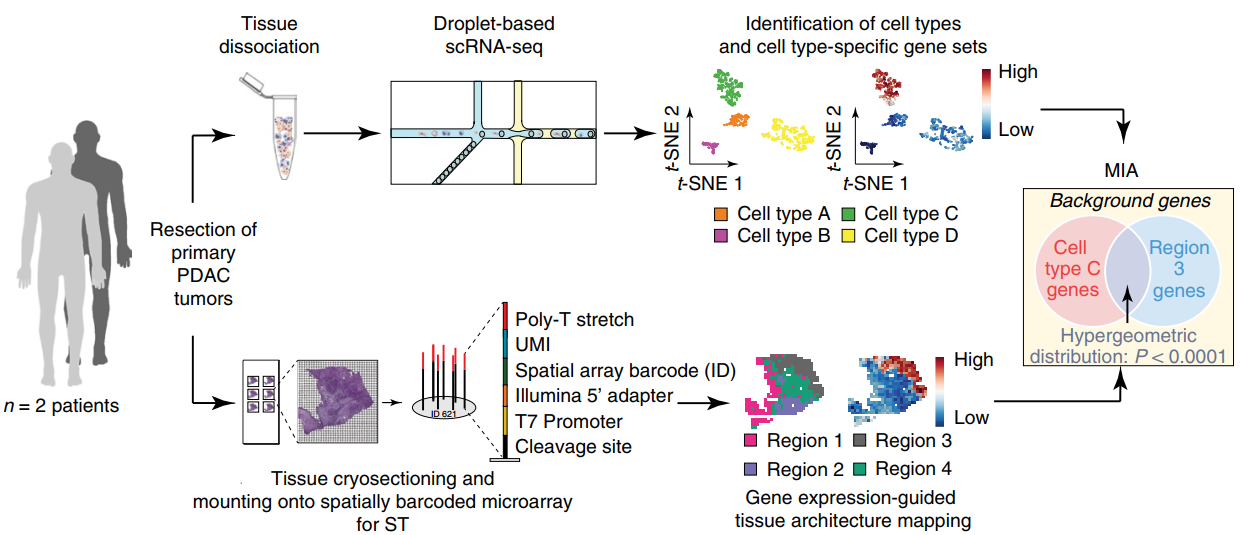
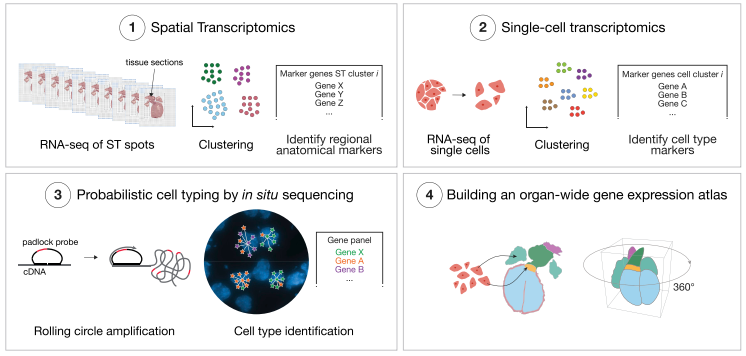
 文献和实验
文献和实验
| 序号 | 伯豪生物提供服务 | 杂志 | IF | 发表年份 |
| 1 | 【样本】:心脏组织 【服务】:空间转录组测序 原文链接【点击查看】 |
Journal of Pharmaceutical Analysis | 8.8 | 2023 |
| 2 | 【样本】:滑膜组织 【服务】:空间转录组测序 原文链接【点击查看】 |
Arthritis and Rheumatology | 13.3 | 2023 |
| 3 | 【样本】:组织 【服务】:样本切片、染色和空转数据分析 原文链接【点击查看】 |
Clinical and Translational Medicine | 8.5 | 2022 |
| 4 | 【样本】:组织 【服务】:空间转录组测序 原文链接【点击查看】 |
Nature Communications | 17.6 | 2022 |
| 5 | ...... |
 技术资料
技术资料
2023公司简介.pdf 附 (下载 1 次)