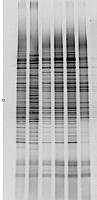
【求助】要做DGGE,真菌有没有通用引物?
丁香园论坛
3842
如题,细菌做DGGE一般是用16S,真菌的有没有,ITS还是18S,GC夹要用什么,加在哪里,上游引物前还是下游引物前?我找过很多文献都没有找到完整的,如果能给出序列(包含GC夹)或者给出相应文献,丁当就奉送。
| FUNGI - UNIVERSAL | |||||
| Name | Region | Fragment Size | Sequence (5' to 3') | Reference | |
| ITS1 | 18S | 570–590 | TCCGTAGGTGAACCTGCGG | White et al. 1990 | |
| ITS4 | 28S | TCCTCCGCTTATTGATATGC | |||
| ITS5 | 18S | 315 | GGAAGTAAAAGTCGTAACAAGG | White et al. 1990 | |
| ITS2 | 5.8S | GCTGCGTTCTTCATCGATGC | |||
| NS1 | 18S | 1100 | GTAGTCATATGCTTGTCTC | White et al. 1990 | |
| NS4 | 18S | CTTCCGTCAATTCCTTTAAG | |||
| NS1 | 18S | 1400 | GTAGTCATATGCTTGTCTC | White et al. 1990 | |
| NS6 | 18S | GCATCACAGACCTGTTATTGCCTC | |||
| NS5 | 18S | 700 | AACTTAAAGGAATTGACGGAAG | White et al. 1990 | |
| NS8 | 18S | TCCGCAGGTTCACCTACGGA | |||
| NS5 | 18S | 310 | AACTTAAAGGAATTGACGGAAG | White et al. 1990 | |
| NS6 | 18S | GCATCACAGACCTGTTATTGCCTC | |||
| White T. J., T. Bruns, S. Lee and J. Taylor, in PCR Protocols: A Guide To Methods And Applications, ed. M. A. Innis, D. H. Gelfand, J. J. Sninsky, T. J. White, Academic Press, San Diego, 1990, pp. 315–322. |
zjubell wrote:
FUNGI - UNIVERSAL Name Region Fragment Size Sequence (5' to 3') Reference ITS1 18S 570–590 TCCGTAGGTGAACCTGCGG White et al. 1990 ITS4 28S TCCTCCGCTTATTGATATGC ITS5 18S 315 GGAAGTAAAAGTCGTAACAAGG White et al. 1990 ITS2 5.8S GCTGCGTTCTTCATCGATGC NS1 18S 1100 GTAGTCATATGCTTGTCTC White et al. 1990 NS4 18S CTTCCGTCAATTCCTTTAAG NS1 18S 1400 GTAGTCATATGCTTGTCTC White et al. 1990 NS6 18S GCATCACAGACCTGTTATTGCCTC NS5 18S 700 AACTTAAAGGAATTGACGGAAG White et al. 1990 NS8 18S TCCGCAGGTTCACCTACGGA NS5 18S 310 AACTTAAAGGAATTGACGGAAG White et al. 1990 NS6 18S GCATCACAGACCTGTTATTGCCTC White T. J., T. Bruns, S. Lee and J. Taylor, in PCR Protocols: A Guide To Methods And Applications, ed. M. A. Innis, D. H. Gelfand, J. J. Sninsky, T. J. White, Academic Press, San Diego, 1990, pp. 315–322.
| BACTERIA - UNIVERSAL | ||||
| Primers | Sequence (5' to 3') | bp | Reference | |
| 63F | 16S | CAG GCC TAA CAC ATG CAA GTC | 1300 | Marchesi et al. 1998 |
| 1387R | GGG CGG WGT GTA CAA GGC | Marchesi et al. 1998 | ||
| 778R | AGG GTA TCT AAT CCT GTT TGC | Rosch & Bothe 2005 | ||
| 27F | 16S | AGA GTT TGA TCM TGG CTC AG | Gurtler & Stanisich 1996 | |
| 1492R | 16S | TAC GGH TAC CTT GTT ACG ACT T | ||
| 338F | Bacteria V3 Region (338-358) | ~undefinedAC TCC TAC GGG AGG CAG CAG | Lane 1991 | |
| 518R | Universal V3 region (534-518) | ATT ACC GCG GCT GCT GG | Muyzer et al. 1993 | |
| 968F | Bacteria V6 region (968-983) | ~undefinedAA CGC GAA GAA CCT TAC | Nübel et al. 1996 | |
| 1406R | Bacteria V9 region (1406-1392) | ACG GGC GGT GTG TAC | Lane et al. 1988 | |
| 72F | 16S | TGC GGC TGG ATC TCC TT | ||
| 38R | 16S | CCG GGT TTC CCCATT CGG | ||
| ~undefinedGC clamp added to the 5' end of the primer, 5'CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G 3' |
附上参考文献,另:丁当不要送我,我拿着没用。
Borneman, J. and R. J. Hartin, Appl. Environ. Microbiol., 2000, 66, 4356.
Braker G, Zhou J, Wu L, Devol AH, Tiedje JM. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096–104
Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775
Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher, J. Clin. Microbiol., 1997, 35, 1353.
Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Gregory, L. G., A. Karakas-Sen, D. J. Richardson, and S. Spiro. 2000. Detection of genes for membrane-bound nitrate reductase in nitrate-respiring bacteria and in community DNA. FEMS Microbiol. Lett. 183:275–279.
Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16
Haynes, K.A., T. J. Westerneng, J. W. Fell and W. Moens, J. Med.Vet. Mycol., 1995, 33, 319.
Lane D.J. 1991. 16S/23S rRNA Sequencing. In: Stackebrandt E., Goodfellow M., eds. Nucleic acid techniques in bacterial systematics. New York: John Wiley & Sons, pp. 115-175.
Lane D.J., Field K.G., Olsen G.J., Pace N.R. 1988. Reverse transcriptase sequencing of ribosomal RNA for phylogenetic analysis. Meth. Enzymol. 167:138-144.
Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiam, D. Dymnock, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799.
Muyzer G., de Waal E.C., Uitterlinden A.G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700.
Muyzer, G., S. Hottentrager, A. Teske, and C. Wawer. 1996. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA-A new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 3.4.4:1–23. In A. Akkermans, et al. (ed.) Molecular microbial ecology manual. Kluwer Academic Publ., Nowell, MA.
Nubel U, B Engelen, A Felske, J Snaidr, A Wieshuber, RI Amann, W Ludwig and H Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriology 178 (19): 5636-5643
Philippot, L., S. Piutti, F. Martin-Laurent, S. Hallet, J.C. Germon. 2002. Molecular Analysis of the Nitrate-Reducing Community from Unplanted and Maize-Planted Soils Appl Environ Microbiol. 2002 December; 68(12): 6121–6128 doi: 10.1128/AEM.68.12.6121-6128.2002.
Redecker D. 2000. Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza 10: 73-80
R?sch, C. & Bothe, H. 2005. Improved Assessment of Denitrifying, N2-Fixing, and Total-Community Bacteria by Terminal Restriction Fragment Length Polymorphism Analysis Using Multiple Restriction Enzymes Applied and Environmental Microbiology, April 2005, p. 2026-2035, Vol. 71, No. 4doi:10.1128/AEM.71.4.2026-2035.2005
R?sch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829
Rotthauwe, J.H., Witzel, K.P. & Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol., 63, 4704–4712.
Sandhu, G. S., B. C. Kline, L. Stockman and G. D. Roberts, J. Clin. Microbiol., 1995, 33, 2931.
Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703
White T. J., T. Bruns, S. Lee and J. Taylor, in PCR Protocols: A Guide To Methods And Applications, ed. M. A. Innis, D. H. Gelfand, J. J. Sninsky, T. J. White, Academic Press, San Diego, 1990, pp. 315–322.
Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554
Borneman, J. and R. J. Hartin, Appl. Environ. Microbiol., 2000, 66, 4356.
Braker G, Zhou J, Wu L, Devol AH, Tiedje JM. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096–104
Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775
Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher, J. Clin. Microbiol., 1997, 35, 1353.
Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Gregory, L. G., A. Karakas-Sen, D. J. Richardson, and S. Spiro. 2000. Detection of genes for membrane-bound nitrate reductase in nitrate-respiring bacteria and in community DNA. FEMS Microbiol. Lett. 183:275–279.
Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16
Haynes, K.A., T. J. Westerneng, J. W. Fell and W. Moens, J. Med.Vet. Mycol., 1995, 33, 319.
Lane D.J. 1991. 16S/23S rRNA Sequencing. In: Stackebrandt E., Goodfellow M., eds. Nucleic acid techniques in bacterial systematics. New York: John Wiley & Sons, pp. 115-175.
Lane D.J., Field K.G., Olsen G.J., Pace N.R. 1988. Reverse transcriptase sequencing of ribosomal RNA for phylogenetic analysis. Meth. Enzymol. 167:138-144.
Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiam, D. Dymnock, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799.
Muyzer G., de Waal E.C., Uitterlinden A.G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700.
Muyzer, G., S. Hottentrager, A. Teske, and C. Wawer. 1996. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA-A new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 3.4.4:1–23. In A. Akkermans, et al. (ed.) Molecular microbial ecology manual. Kluwer Academic Publ., Nowell, MA.
Nubel U, B Engelen, A Felske, J Snaidr, A Wieshuber, RI Amann, W Ludwig and H Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriology 178 (19): 5636-5643
Philippot, L., S. Piutti, F. Martin-Laurent, S. Hallet, J.C. Germon. 2002. Molecular Analysis of the Nitrate-Reducing Community from Unplanted and Maize-Planted Soils Appl Environ Microbiol. 2002 December; 68(12): 6121–6128 doi: 10.1128/AEM.68.12.6121-6128.2002.
Redecker D. 2000. Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza 10: 73-80
R?sch, C. & Bothe, H. 2005. Improved Assessment of Denitrifying, N2-Fixing, and Total-Community Bacteria by Terminal Restriction Fragment Length Polymorphism Analysis Using Multiple Restriction Enzymes Applied and Environmental Microbiology, April 2005, p. 2026-2035, Vol. 71, No. 4doi:10.1128/AEM.71.4.2026-2035.2005
R?sch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829
Rotthauwe, J.H., Witzel, K.P. & Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol., 63, 4704–4712.
Sandhu, G. S., B. C. Kline, L. Stockman and G. D. Roberts, J. Clin. Microbiol., 1995, 33, 2931.
Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703
White T. J., T. Bruns, S. Lee and J. Taylor, in PCR Protocols: A Guide To Methods And Applications, ed. M. A. Innis, D. H. Gelfand, J. J. Sninsky, T. J. White, Academic Press, San Diego, 1990, pp. 315–322.
Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554
zjubell wrote:
你搜索一下看,这篇文献是非常经典的。呵呵。同样,再给出一个细菌的。
BACTERIA - UNIVERSAL Primers Sequence (5' to 3') bp Reference 63F 16S CAG GCC TAA CAC ATG CAA GTC 1300 Marchesi et al. 1998 1387R GGG CGG WGT GTA CAA GGC Marchesi et al. 1998 778R AGG GTA TCT AAT CCT GTT TGC Rosch & Bothe 2005 27F 16S AGA GTT TGA TCM TGG CTC AG Gurtler & Stanisich 1996 1492R 16S TAC GGH TAC CTT GTT ACG ACT T 338F Bacteria V3 Region (338-358) ~undefinedAC TCC TAC GGG AGG CAG CAG Lane 1991 518R Universal V3 region (534-518) ATT ACC GCG GCT GCT GG Muyzer et al. 1993 968F Bacteria V6 region (968-983) ~undefinedAA CGC GAA GAA CCT TAC Nübel et al. 1996 1406R Bacteria V9 region (1406-1392) ACG GGC GGT GTG TAC Lane et al. 1988 72F 16S TGC GGC TGG ATC TCC TT 38R 16S CCG GGT TTC CCCATT CGG ~undefinedGC clamp added to the 5' end of the primer, 5'CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G 3'
谢谢!
本文由丁香园论坛提供,想了解更多有用的、有意思的前沿资讯以及酷炫的实验方法的你,都可以成为师兄的好伙伴
师兄微信号:shixiongcoming