Analyzing Transcription Factor Occupancy During Embryo Development Using ChIP-seq
互联网
互联网
相关产品推荐

Recombinant-Arabidopsis-thaliana-Sterol-14-demethylaseCYP51G1Sterol 14-demethylase EC= 1.14.13.70 Alternative name(s): Cytochrome P450 51A2 Cytochrome P450 51G1; AtCYP51 Obtusifoliol 14-demethylase Protein EMBRYO DEFECTIVE 1738
¥13118
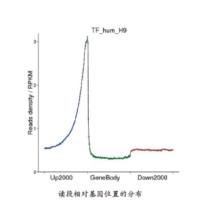
ChIP-seq
询价
促销中大鼠肿瘤坏死因子α(TNF-α)/Rat TNF-α/tumor necrosis factor (TNF superfamily,member 2)/Tnf;Tnfa;Tnfsf2;Tumor necrosis factor;Cachectin;TNF-alpha;Tumor necrosis factor ligand superfamily member 2;TNF-a) [Cleaved into: Tumor necrosis factor;membrane for/TNF/ELISA试剂盒
¥3420¥3800

Recombinant-Arabidopsis-thaliana-Protein-TIC-40-chloroplasticTIC40Protein TIC 40, chloroplastic Alternative name(s): Protein PIGMENT DEFECTIVE EMBRYO 120 Translocon at the inner envelope membrane of chloroplasts 40; AtTIC40
¥12082

Recombinant-Arabidopsis-thaliana-Mitochondrial-import-inner-membrane-translocase-subunit-TIM50TIM50Mitochondrial import inner membrane translocase subunit TIM50 Alternative name(s): Protein EMBRYO DEFECTIVE 1860
¥11900
相关问答

