The Use of RAPD for Isolate Identification of Arbuscular Mycorrhizal Fungi
互联网
587
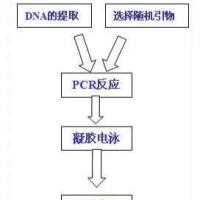
The basic polymerase chain reaction (PCR) ( 1 ), which consists of the exponential amplification of DNA fragments between terminal sequences recognized by specific oligonucleotide primers, is a very powerful method but does not satisfy all wishes. The need for sequence information from the DNA template, in order to create the primers, limits the applicability of the method to a few organisms or to highly conserved genomic DNA regions. This is a handicap if highly variable regions need to be analyzed, as is normally the case for the identification of organisms at the strain or isolate level. To circumvent this problem, the basic PCR method has been modified. Instead of long oligonucleotide (20–30 mer) primers, short (9–13 mer) oligonucleotides of arbitrary sequence can be used. They bind to complementary sequences occurring more or less frequently in the genome. A DNA amplification product is generated for each genomic region that happens to be flanked by a pair of priming sites that are within 3000 bp of each other and in the appropriate orientation (Fig. 1 ).
Fig. 1. Amplification of genomic DNA with short arbitrary primers. Fragments, which are flanked by a pan of priming sites within 3000 basepans in the appropriate orientatton (A), are amplified exponentially (A′). Fragments with one priming site (B) or with more than 3000 bp within a pan of priming sites (C) are not amplified exponentially (B′, C′ ).