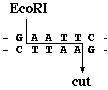RFLP Method
互联网
- 相关专题
RFLP (often pronounced "rif lip", as if it were a word) is a method used by molecular biologists to follow a particular sequence of DNA as it is passed on to other Cell s. RFLPs can be used in many different settings to accomplish different objectives. RFLPs can be used in paternity cases or criminal cases to determine the source of a DNA sample. RFLPs can be used determine the disease status of an individual. RFLPs can be used to measure recombination rates which can lead to a genetic map with the distance between RFLP loci measured in centiMorgans.
On this web page, you can see how RFLPs are produced and then three examples of applying RFLP analysis: paternity , disease status , and genetic mapping .
RFLP Production
Each organism inherits its DNA from its parents. Since DNA is replicated with each generation, any given sequence can be passed on to the next generation. An RFLP is a sequence of DNA that has a restriction site on each end with a "target " sequence in between. A target sequence is any segment of DNA that bind to a probe by forming complementary base pairs. A probe is a sequence of single-stranded DNA that has been tagged with radioactivity or an enzyme so that the probe can be detected. When a probe base pairs to its target, the investigator can detect this binding and know where the target sequence is since the probe is detectable. RFLP produces a series of bands when a Southern blot is performed with a particular combination of restriction enzyme and probe sequence.
For example, let"s follow a particular RFLP that is defined by the restriction enzyme EcoR I and the target sequence of 20 bases GCATGCATGCATGCATGCAT. EcoR I binds to its recognition seuqence GAATTC and cuts the double-stranded DNA as shown:

In the segement of DNA shown below, you can see the elements of an RFLP; a target sequence flanked by a pair of restriction sites. When this segment of DNA is cut by EcoR I, three restriction fragments are produced, but only one contains the target sequence which can be bound by the complementary probe sequence (purple ).

Let"s look at two people and the segments of DNA they carry that contain this RFLP (for clarity, we will only see one of the two stands of DNA). Since Jack and Jill are both diploid organisms, they have two copies of this RFLP. When we examine one copy from Jack and one copy from Jill, we see that they are identical:
Jack 1: -GAATTC---(8.2 kb)---GCATGCATGCATGCATGCAT ---(4.2 kb)---GAATTC-
Jill 1: -GAATTC---(8.2 kb)---GCATGCATGCATGCATGCAT ---(4.2 kb)---GAATTC-
When we examine their second copies of this RFLP, we see that they are not identical. Jack 2 lacks an EcoR I restriction site that Jill has 1.2 kb upstream of the target sequence (difference in italics).
Jack 2: -GAATTC--(1.8 kb)-CCCTTT --(1.2 kb)--GCATGCATGCATGCATGCAT --(1.3 kb)-GAATTC-
Jill 2: -GAATTC--(1.8 kb)-GAATTC --(1.2 kb)--GCATGCATGCATGCATGCAT --(1.3 kb)-GAATTC-
Therefore, when Jack and Jill have their DNA subject to RFLP analysis, they will have one band in common and one band that does not match the other"s in molecular weight: