“多莉”的诞生,意味着人类可以利用动物的一个组织细胞 ,像翻录磁带或复印文件一样,大量生产出相同的生命体,这无疑是基因工程研究领域的一大突破。 人们剪下植物枝条,扦插到土里,不久就会发芽,长出新的植株,这些植株是遗传物质组成完全相同的植株,这就是“克隆 ”。还有将马铃薯等植物的块茎切成许多小块进行繁殖,由此而长出的后代也是“克隆 &rd ...
一、DNA 提取 SDS 高盐法(方案1) 具体步骤: 称取1 g 土壤,放入研钵中,倒入适量的液氮,立即研磨;再倒入适量的液氮,研磨,重复3 次,使土壤颗粒研成粉末; 将13.5 ml 提取缓冲液(0.1 mol/L磷酸盐 ,0.1 mol/L EDTA ,0.1 mol/L Tris-HCl ,1.5 mol/L NaCl ,1.0 % CTAB) 和50&mu ...
序列测定的技术和策略 Sanger双脱氧链终止法 Maxam-Gilbert DNA 化学降解法 测序策略 目前应用的两种快速序列测定技术是Sanger等(1977)提出的酶法及Maxam和Gilbert(1977)提出的化学降解法。虽然其原理大相径庭,但这两种方法都是同样生成互相独立的若干组带放射性标记的寡核苷酸,每组寡核苷酸都有固定的起点,但却随机终止于特定的一种或者多种残基上。由于DNA 上 ...
转染的定义是“将具生物功能的核酸转移或运送到细胞内并使核酸在细胞内维持其生物功能”。其中,核酸包括DNA (质粒和线性双链DNA ),反义寡核苷酸及RNAi(RNA interference)。基因转染技术已广泛应用于基因组功能研究(基因表达调控,基因功能,信号转导和药物筛选研究)和基因治疗研究。基因转染需要一定的转染试剂将带有目的基因的载体运送到细胞内。早期的磷酸钙转染法转染效率很低,且对很多细胞株无效,因此不能满足很多科 ...
alimama_pid="mm_10043573_137377_1822161"; alimama_titlecolor="FF9900"; alimama_descolor ="552c55"; alimama_bgcolor="f7f7f7"; alimama_bordercolor="f7f ...
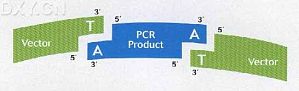
PCR克隆主要有TA克隆法 限制性酶切与连接法,杂交法和近期开发出来的特异性重组法等。 但因为TA克隆法操作最简单,快速和高效的原因,成为Taq聚合酶PCR产物的最佳克隆方法。TA克隆法由Invitrogen发明,并拥有全球TA Cloning商标的专利权。 TA克隆方法(Original TA Cloning Kit) 利用Taq聚合酶同时具有的末端连接酶的功能,在每条PCR扩增产物的3`端自动 ...
第一节 概 述 质粒具有稳定可靠和操作简便的优点。如果要克隆 较小的DNA 片段(<10kb)且结构简单质粒要比其它任何载体都要好。在质粒载体上进行克隆 从原理上说是很简单的,先用限制性内切酶切割质粒DNA 和目的DNA 片段 然后体外使两者相连接 再用所得到重组质粒转化细菌即可完成。但在实际工作中 如何区分插入有外源DNA 的重组质粒和无插入而自身环化的载体分子是较为困难的。通过调整连接反应中 ...
alimama_pid="mm_10043573_137377_1822161"; alimama_titlecolor="FF9900"; alimama_descolor ="552c55"; alimama_bgcolor="f7f7f7"; alimama_bordercolor="f7f7f7"; alimama_linkcolor="552c55"; ...
体外定点突变技术是研究蛋白质结构和功能之间的复杂关系的有力工具,也是我们在实验室中改造/优化基因常用的手段。蛋白质的结构决定其功能,二者之间的关系是蛋白质组 研究的重点之一。对某个已知基因的特定碱基进行定点改变、缺失或者插入,可以改变对应的氨基酸序列和蛋白质结构,对突变基因的表达产物进行研究有助于我们了解蛋白质结构和功能的关系,探讨蛋白质的结构/结构域。而利用定点突变技术改造基因,相信大家也非常熟 ...
在分子生物学研究中,DNA 的序列分析是进一步研究和改造目的基因的基础。目前用于测序的技术主要有Sanger等(1977)发明的双脱氧链末端终止法和Maxam和 Gilbert(1977)发明的化学降解法。这二种方法在原理上差异很大,但都是根据核苷酸在某一固定的点开始,随机在某一个特定的碱基处终止,产生A,T,C,G四组不同长度的一系列核苷酸,然后在尿素变性的PAGE胶上电泳进行检测,从而获得DN ...
第一节 概 述 DNA分子水平上的多态性检测技术是进行基因组研究的基础。RFLP(Restriction Fragment Length Polymorphism限制片段长度多态性)已被广泛用于基因组遗传图谱构建、基因定位以及生物进化和分类的研究。RFLP是根据不同品种(个体)基因组的限制性内切酶的酶切位点碱基发生突变,或酶切位点之间发生了碱基的插入、缺失,导致酶切片段大小发生了变化,这种变化可以 ...
当双链DNA 分子的一条链上产生切口时,E.coli DNA 聚合酶Ⅰ就可将核苷酸连接到切口的3'羟基末端。同时该酶具有从5'→3'的核酸外切酶活性,能从切口的5'端除去核苷酸。由于在切去核苷酸的同时又在切口的3'端补上核苷酸,从而使切口沿着DNA 链移动,用放射性核苷酸代替原先无放射性的核苷酸,将放射性同位素掺入到合成新链中。最合适的切口平移片段一般为50-500个核苷酸。切口平移反应受几种因素的影响: (a) 产物的比活性取决于dNTP ...
DNA 是遗传信息的载体,是最重要的生物信息分子,是分子生物学研究的主要对象,因此DNA 的提取也应是分子生物学实验技术中的最重要、最基本的操作,如不能有效的完成DNA 提取方面的工作,那就根本谈不上进行分子生物学方面的实验。在DNA 提取过程中应做到1,根据不同研究需要,保证结构的相应完整性;2,尽量排除其它大分子成分的污染(蛋白质、多糖及RNA 等);3,保证提取样品中不含对酶有抑制作用的有机 ...
无论是从一个胚胎细胞分化为不同的组织器官,或者是从正常的组织突变为肿瘤组织,都可能涉及在同一基因组 背景下不同基因的差异性表达。寻找差异表达的基因就有可能揭示细胞分化的机制或者肿瘤的成因,因而也就成为一项热门的技术。 寻找差异表达的基因有不少方法,抑制消减杂交(SSH)是比较有效的一种方法。以Clontech的PCR-Select cDNA Subtraction Kit为例:将样品(teste ...
一、 建立一个标准的酶切反应目前大多数研究者遵循一条规则,即10个单位的内切酶可以切割1μg不同来源和纯度的DNA。通常,一个50μl的反应体系中,1μl的酶在1X NEBuffer终浓度及相应温度条件下反应1小时即可降解1μg已纯化好的DNA。如果加入更多的酶,则可相应缩短反应时间;如果减少酶的用量,对许多酶来说,相应延长反应时间(不超过16小时)也可完全反应。二、 选择正确的酶不言而喻,选择的酶在底物DNA上必须至少有 ...
一、核酸的纯化 在分子克隆 的所有操作中,最基本的操作是核酸的纯化。其关键步骤是去蛋白质,通常只要用酚/氯仿。氯仿抽提核酸的溶液即可。每当需要把克隆 有某一些所用的酶灭活或去除以便进行下一步时,可进行这种抽提。然而,如要从细胞裂解液等复杂的分子混合物中纯化核酸,则要先用某些蛋白水解酶消化大部分蛋白质后,再用有机溶剂抽提。这些广谱的蛋白酶包括链霉蛋白酶及蛋白酶K等,它们对多种天然蛋白均有活性,(1) ...
Marian Price-Carter, 9/7/00Day 1 : Start overnight cultures in assay medium.Negative control : cells lacking b -galactosidase, such as LT2; positive control : cells with high enzyme activity.Day 2 : Dilute cells 1/100 in fresh medium, grow to mid-log.1 Prepare solutions: Z buffer, phosphate buf ...
Materials: phenol:chloroform (1:1) chloroform Add an equal volume of buffer-saturated phenol:chloroform (1:1) to the DNA solution. Mix well. Most DNA solutions can be vortexed for 10 sec except for high molecular weight DNA which should be gently rocked. (If using Phase-Lock Gel, follow procedure M.1 ) Spin in a microfuge for 3 min. Carefully remove the aqueous layer to a new tube, being careful to avoid the interface. (Steps 1-4 can be repeated until an interface is no longer visible) To
Footprinting Footprinting is a method for determining the exact DNA sequence to which a particular DNA-binding protein binds. Examples: hormone-receptor complexes that bind to their hormone response elements transcription factors that bind eukaryotic operators, enhancers, and silencers the lac repressor that shuts down the lac operon in E. coli [Discussion of the lac operon] How, for example, does one determine the DNA sequence to which the lac repressor binds? The procedure: Clon
alimama_pid="mm_10043573_137377_1822161"; alimama_titlecolor="FF9900"; alimama_descolor ="552c55"; alimama_bgcolor="f7f7f7"; alimama_bordercolor="f7f ...