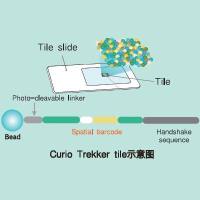
Low-Resolution Mapping of Untagged Mutations
互联网
521
The mapping method detailed here is based on the multiplex polymerase chain reaction (PCR) coamplification of 32 molecular markers, using fluorescently labeled oligonucleotides as primers. For the genotyping of a single plant from a mapping population, only two simultaneous amplifications are required, the products of which are finally electrophoresed in an automated DNA sequencer controlled by fragment analysis software. An analysis of the genotypes of 50 plants allows mapping of the mutation of interest within a candidate genomic interval of about 15 cM (3 Mb, corresponding to about 40 BAC clones).