Next-Generation Sequencing of miRNAs with Roche 454 GS-FLX Technology: Steps for a Successful Application
互联网
865
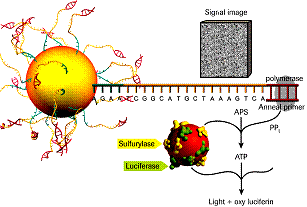
MicroRNAs (miRNAs) are a class of small RNAs (sRNAs) of approximately 22 nucleotides in length that control eukaryotic gene expression at the translational level. They regulate a wide variety of biological processes, namely developmental timing, cell differentiation, cell proliferation, the immune response, and infection. Their identification is essential to understand eukaryotic biology. Their small size, low abundance, and high instability complicated early identification, however new generation genome sequencing approaches, such as the Roche 454 Pyrosequencer, allow for both miRNA identification and for generating miRNA profiles in a given sample. This technique avoids cloning steps in bacteria and is a fast and bias-minimized tool to discover novel miRNAs and other sRNAs on a genome-wide scale. Prior to sequencing, cDNA libraries are built for each sample using total RNA as starter material. Each cDNA library can be tagged with specific identifier sequences that allow sequencing different samples in the same chip run. Here, we describe the protocols for the construction of sRNA cDNA libraries for 454 sequencing, and we include tips for overcoming problems often encountered during cDNA library preparation.