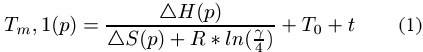Real-Time Primer Design for DNA Chips
互联网
|
|
H. Simmler and H. Singpiel
Acconovis GmbH
Lindenhofstr. 42-44
68163 Mannheim, Germany
eMail: simmler@acconovis.com
R. M¨anner
Universit¨at Mannheim
B6, 23-29
68131 Mannheim, Germany
maenner@ti.uni-mannheim.de
Abstract
The design of PCR or DNA Chip experiments is a time consuming process where bioinformatics is extensively used. The selection of the primers, which are immobilized on the DNA chip, requires a complex algorithm. Based on several parameters an optimized set of primers is automatically detected for a given gene sequence.This paper describes a parallel architecture which performs the optimization of the primer selection on a hardware accelerator. In contrast to the pure software approach,the parallel architecture gains a speedup of factor 500 using a PCI based hardware accelerator. This approach allows an optimization of a specified primer set in real-time.
1 Introduction
Both, the amplification of DNA sequences using polymerase chain reaction (PCR) and the massive parallel analysis
of genes in biological cells using DNA chips (or DNA arrays) have a great impact on modern biological research. PCR is used to amplify a particular DNA fragment called target sequence. In general, a forward and a reverse primer is generated. The target sequence, located between the two primers, is duplicated using a complex process protocol