E.Z.N.A.® Total RNA Midi Kit Protocol for Eukaryotic Cells and Tissues
互联网
922
互联网
相关产品推荐
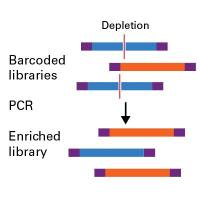
链特异性RNA建库试剂SMART-Seq® Total RNA Pico Input with UMIs (ZapR™ Mammalian)
¥18658

免疫沉淀protocol
¥600

BEAS-2B人正常肺上皮细胞|BEAS-2B细胞系|Human Normal Lung Epithelial Cells
¥1500
E.Z.N.A.™ BAC/PAC DNA Kit
¥550

RELT/RELT蛋白Recombinant Human Tumor necrosis factor receptor superfamily member 19L protein (RELT)重组蛋白Receptor expressed in lymphoid tissues蛋白
¥1344
相关问答

