Creating a deletion by PCR spli
互联网
Creating a deletion by PCR splicing
Contributed by Dr.A.Gratchev
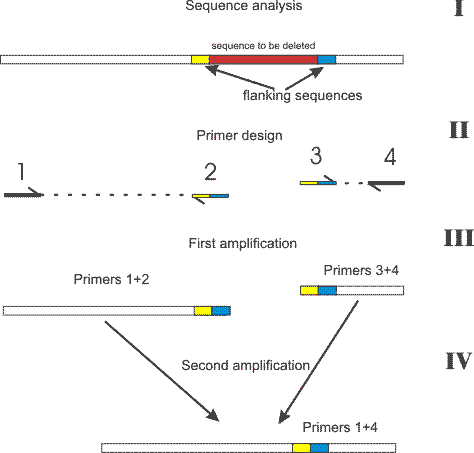
I didn't want to place this in the methods section due to its simplicity. To delete a desired fragment from existing DNA fragment all you need is a pair of primers combined of flanking sequences and a high fidelity polymerase (a mix of Taq and Pfu for example). The procedure is shown on the picture. The only notes which I want to make are:
- If you use the plasmid as a template use about 500 ng. In this case you can make only 20 cycles to have a good product.
- Primers 1 and 4 can be any, primers 2 and 3 should be about 30 bases (15 per flank), but you can anyway use annealing at 55°C.
- No purification of first stage product needed, just dilute them 1:100 to reduce the amount of the primers 2 and 3 which you don't need any more.
- The same method can be used to create point mutations.
上一篇:In vitro site-specific mutagenesis 下一篇:Erase-a-Base® System