ABSTRACTThe simplest way to determine whether two monoclonal antibodies bind to distinct sites on a protein antigen is to carry out a competition assay. The assay can be used with antibodies that bind ...
INTRODUCTIONOne versatile strategy for sample cleanup prior to MALDI-MS analysis uses microscale columns designed for direct sample elution onto the MALDI target plate. This protocol describes the fab ...
TILLING in the Botanical Garden final A17 published version 06.pdfTarget-selected mutant screen by TILLING in Drosophila.pdfTILLING (Targetting Induced Local Lesions In Genomes) Technology for Plant F ...
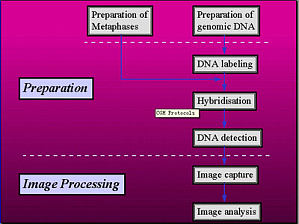
Theory of CGHComparative genomic hybridization (CGH) is a fairly new molecular cytogenetic technique that allows detection of DNA sequence copy number changes throughout the genome in a single hybridi ...
Metaphase chromosome preparation Materials: RPMI 1640 medium fetal calf serum (FCS) 20% Colcemid (e.g. Boehringer Mannheim cell biology reagents Best.-Nr. 295892) cell cuture flask Phythemaglutinin PH ...
DNA preparation by cryotom tissue dissection Preparations/Materials: Cool cryostat down to -20 to -30°C about 3 hours prior to dissection Label eppendorf tube (2 ml e.g. Safe Lock) and microscopic sli ...
Hybridization reagents: labeled tumor and normal-DNA (see protocol Nick translation) salmon sperm DNA 10 mg/ml (e.g. Promega) human Cot1 DNA 1 mg/ml (GibcoBRL Life Technologies) 3M sodium acetate 100% ...
CGH Image acquisition Images were acquired through a Zeiss Axiophot fluorescence microscope using a Plan NEOFLUAR oil objective x63 N.A. 1.25 (Zeiss Oberkochen Germany) equipped with filter sets appro ...
Comparative genomic hybridization (CGH) is a molecular cytogenetic method for the detection of chromosomal imbalances. In a CGH experiment two genomic DNA samples are simultaneously hybridized in sit ...
This CGH Protocol is used for DNA of good quality when available in sufficient amounts. We usually do replicate hybridizations using samples labeled "inversely" (reversing the label for test and norma ...
DAY 1:1) Reprecipitate DNA'sAdd the following DNA's to a 1.5 ml centrifuge tube mixing with pipet:20 ug Cot-1 DNA (~20ul)~200 ng Biotin labeled DNA (~10ul)~200 ng Digoxigenin labeled DNA (~10ul) Add 1 ...
Random Prime Amplification of DNA from small tumors (fresh or paraffin)We generally use 50 nanograms of fresh or paraffin microdissected Taqman-quantitated DNA (see microdissection protocol) for each ...
Cut 10-20X 10 um sections of formalin fixed paraffin samples into eppendorf tubes. Add 1 ml xylene mix incubate at 55 C for 15 mins. Release pressure spin down for 2 minutes in eppendorf ultramicrofug ...
In designing the PCR reaction be sure to include a negative PCR control (blank without DNA) a positive control (we use MPE-600 breast tumor cell line DNA) and enough reference DNA for all of the plann ...
Tissue Handling: Note that all unfixed human tissue should be handled as BioSafety Level 2 materials (wear gloves lab coat etc). All plasticware (tubes tips etc.) should be RNase free and always handl ...
1. Prepare reaction mixtures per 50ul Add Enzymes last gently vortex mixture quick spin liquid to bottom of tube: LabelBiotin-dUTP Digox-dUTP FITC-dUTP Texas Red dUTP 10X Biotin dNTP5 ul 0 ul 0 ul 0 u ...
IntroductionThe technique of chromatin immunoprecipitation (ChIP) has proven to be a powerful tool allowing the detection of protein-DNA interactions in living cells. Hybridization of ChIP samples to ...
Comparative genomics has become a real tantalizing challenge in the postgenomic era. This fact has been mostly magnified by the plethora of new genomes becoming available in a daily bases. The overwhe ...
Gene Set Analysis and Network Analysis for Genome-Wide Association StudiesInti Pedroso and Gerome Breen INTRODUCTIONThe application of high-throughput genotyping in humans has yielded numerous insight ...
The software included are listed bellowIMPUTEa program for genotype imputation in genome-wide association studies and fine-mapping studies based on a dense set of marker data (such as HapMap) 5v1.0.0 ...