CGH Protocols (二)
互联网
DNA preparation by cryotom tissue dissection
Preparations/Materials:
Cool cryostat down to -20 to -30°C about 3 hours prior to dissection
Label eppendorf tube (2 ml, e.g. Safe Lock) and microscopic slides with the case number
Digestion buffer (50 ml Tris, pH 8.5, 1 mM EDTA, 0.5% Tween 20)
Proteinase K (500 µl aliquots of a 20mg/ml stock solution, keep aliquots at -20°C)
Melt the tip of long Pasteur pipettes (one pipette per case)
Dissect only one block per time in the cryostat
Keep other tissue block frozen (e.g. in styropor box with liquid nitrogen or dry ice) during dissection
Take care to replace tissue block in the correct vial after dissection

Steps:
- Freeze tissue block (0.5 - 1 cm2 ) on the cryostat plate, e.g. by applying few drops of isotonic NaCl or water and placing the tissue immediately on the cooled cryostat plate
- Cut the tissue block and place first section (5-8 µm) on labeled glass slide (close to case No. label)
- Cut about 20-30 section (20-30µm) and transfer them in the labeled eppendorf tube which contains 900µl digestion buffer. The transfer is facilitated by picking the cool sections by the tip of the pasteur pipette. Change pipette after each case!
- Transfer last section (5-8 µm) on the glass slide (distal to case No. label)
- Add 30-50 µl of Proteinase K (stock solution of 20 mg/ml)
- Incubate for at least 2 hours at 50°C, check the digestion by the disintegration if the tissue, add new Proteinase K if the digestion is bad. The Proteinase K digestion can be extended over night or even for several days.
- Add 1000 µl of Phenol/Chloroform/Isoamylalcohol, mix e.g. by inverting for about 10 min, centrifuge in a table top centrifuge for 10-20 min until the two phases have clearly separated. Discard upper phase.
- Repeat step 7 at least once (better: twice).
- Add 1/10 vol (about 90 µl) 3 M NaCl, mix. Add 1 vol (about 1000 µl) ice cool isopropanol. Mix by inverting (white DNA pellet visible? If yes, DNA amount and quality usually oK for Nick translation and CGH)
- Centrifuge DNA pellet for about 5 min, discard supernatant, wash once with 100% EtOH and once with 70% EtOH. Finally air dry pellet or dry it by a SpeedVac (5-10 min). If you use a SpeedVac take care not to dry the DNA too much since high molecular, ultradry DNA may be difficult to dissolve
- Dissolve pellet in about 100-200µl H2 0 (e.g. Acqua ad iniectabile, Braun Melsungen) depending on the amount of DNA
- Determine DNA concentration by a photometer (Final concentration for nick translation should by higher than 150 µg/ml).
- Check amount of tumor DNA in the tissue block by an H&E stain of the slide (Amount of tumor DNA compared to normal stromal tissue should exceed 70%)
DNA labeling by nick translation
reagents:
DNA for labeling (concentration c > 150 ng/µl)
modified nucleotides: Bio tin-16-dUTP, Dig oxigenin-11-dUTP, conc. 1nmol/µl (Boehringer Mannheim)
dNTPs (regular nucleotides): dATP, dCTP, dGTP, 0.5 mM each, dTTP 0.1 mM
NT reaction buffer 10x (0.5 M Tris pH 8, 50 mM MgCl 2 , 0.5mg/ml BSA)
b -ME (beta-mercaptoethanol) 0.1 M
DNase (stock solution 3 mg/ml) 1:2000 diluted in aqua bidest.
Pol: Kornberg DNA-polymerase 5 U/µl (e.g. Boehringer Mannheim)
EDTA (0.5 M, pH 8.0)
SDS (20%)
for one NT reaction 5 µg of DNA is used:
| Mix (V total = 50 µl): | 1 probe | mix for N probes | |
| NT (10x) | 5 µl | (N+1) * 5 : | |
| b -ME | 5 µl | (N+1) * 5 : | for more than 1 probe |
| dNTPs | 5 µl | (N+1) * 5 : | pipette 19 µl to the |
| Bio/Dig-dUTundefined | 2 µl | (N+1) * 2 : | DNA+H 2 O |
| DNase (1:2000) | 1 µl | (N+1) * 1 : | |
| Pol | 1 µl | (N+1) * 1 : | |
| --------------------- | --------------------- | --------------------- | |
| DNA+H 2 O | 31 µl | ||
| ===== | |||
| 50 µl |
Pipette on ice!
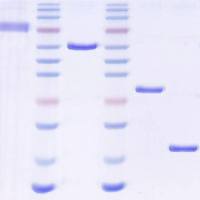
incubation for 2 hrs at 15°C --> put probes on ice --> test 5 µl of the mix in an agarose electrophoresis, optimal length of DNA fragments should be between 100-1000 bp (in the mean time store probes at -20°C)
-->if neccessary incubate longer after addition of new DNAse and Pol
-->add 2.5 µl EDTA (0.5 M, pH 8.0) and 2.5 µl SDS (20%) to stop the reaction, keep the probes at -20°C until hybridization
Optimal fragment length after nick translation
| DNA after agarose gel | ===> | Detection of labeled DNA by a color reaction |
| electrophoresis | after transfer to a nylon membrane |