相关产品推荐更多 >
万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 供应商:
环亚生物科技有限公司
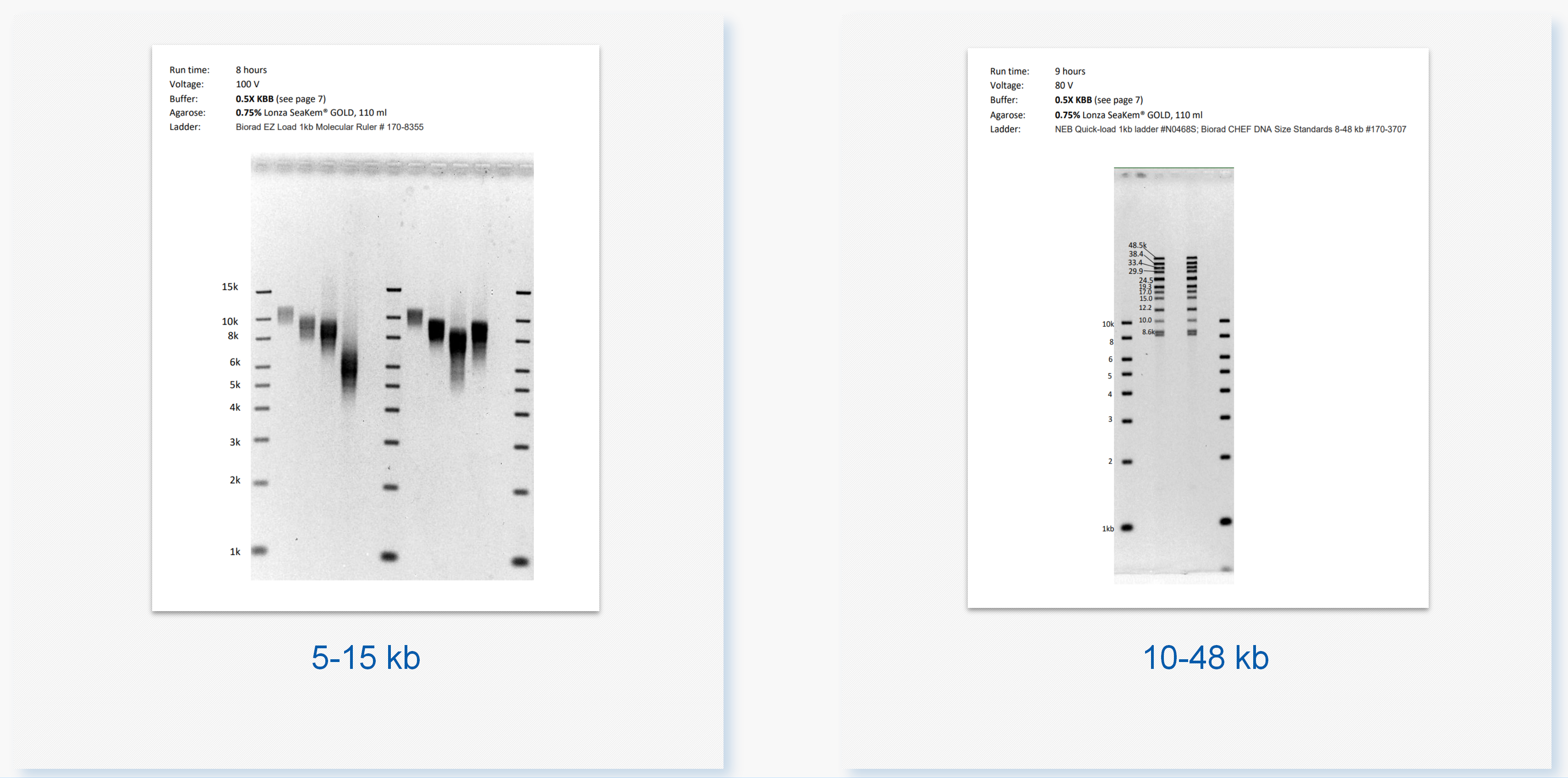
Pippin Pulse—脉冲场电泳仪,用于基因组大片段DNA的电泳分离检测:分离0.5kb-430kb
• Sage Science 公司
位于美国马萨诸塞州,2010年成立,专注于核酸领域的样品制备
全球超过3000家客户的认可,国内装机数量400台左右
2万篇文献中应用,包括众多Nature、 Science、 Cell系列的文章
DNA片段筛选回收领域的行业金标准
• Pippin Pulse 脉冲场电泳仪
通过脉冲场电泳,分离DNA大片段
用于DNA大片段的分离检测和质控
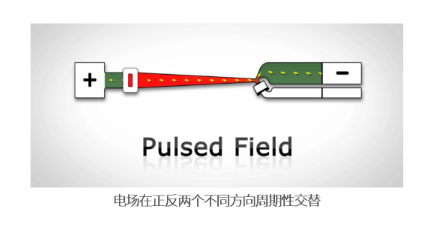
技术原理
脉冲场电泳 (Pulse Field Gel Electrophoresis, PFGE) 原理:
- 电场不是恒定的,是在正反两个不同方向间进行周期性交替
- 在交替变换方向的电场中,DNA分子作出反应、调整构象所需的时间
- 分离迁移的速度依赖于分子大小
产品特点
- 自带经过验证的多种程序。
- 支持自定义程序
- 加固的大直径铂电极,寿命更长
- 试剂耗材开放
应用方向

典型案例
|
案例1:预设程序
|
|
案例2:Pippin Pulse + Nanopore
|
|
案例3:Pippin Pulse + Pacbio
|
|
案例4:Pippin Pulse + Bionano
|
代表客户

风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验发表文献
Pippin Pulse + Nanopore:
|
1.Long, Meng, et al. "Comparative genomic analysis provides insights into taxonomy and temperature adaption of Aeromonas salmonicida." Journal of Fish Diseases 46.5 (2023): 545-561. 2.Keraite, Ieva, et al. "A method for multiplexed full-length single-molecule sequencing of the human mitochondrial genome." Nature Communications 13.1 (2022): 5902. 3.Ong, Chian Teng, et al. "Adaptive sampling during sequencing reveals the origins of the bovine reproductive tract microbiome across reproductive stages and sexes." Scientific reports 12.1 (2022): 15075. 4.Wunderlich, Stephanie, et al. "Targeted biallelic integration of an inducible Caspase 9 suicide gene in iPSCs for safer therapies." Molecular Therapy-Methods & Clinical Development 26 (2022): 84-94. 5.Russo, Alessia, et al. "Low-input high-molecular-weight DNA extraction for long-read sequencing from plants of diverse families." Frontiers in Plant Science 13 (2022): 1494. |
Pippin Pulse + PacBio:
|
1.Takeuchi, Tadashi, et al. "Fatty acid overproduction by gut commensal microbiota exacerbates obesity." Cell Metabolism (2023). 2.Hadzhiev, Yavor, et al. "The miR-430 locus with extreme promoter density forms a transcription body during the minor wave of zygotic genome activation." Developmental Cell 58.2 (2023): 155-170. 3.Kurokochi, Hiroyuki, et al. "Telomere-to-telomere genome assembly of matsutake (Tricholoma matsutake)." DNA Research 30.3 (2023): dsad006. 4.Hixson, Kim K., et al. "Annotated genome sequence of a fast-growing diploid clone of red alder (Alnus rubra Bong.)." G3: Genes, Genomes, Genetics (2023): jkad060. 5.Field, Matt A., et al. "The Australian dingo is an early offshoot of modern breed dogs." Science Advances (2022): eabm5944. |
Pippin Pulse + Bionano:
|
1.Secomandi, Simona, et al. "A chromosome-level reference genome and pangenome for barn swallow population genomics." Cell Reports 42.1 (2023): 111992. 2.Dahn, Hollis A., et al. "Benchmarking ultra-high molecular weight DNA preservation methods for long-read and long-range sequencing." GigaScience 11 (2022). 3.Palmada-Flores M, Orkin J D, Haase B, et al. A high-quality, long-read genome assembly of the endangered ring-tailed lemur (Lemur catta)[J]. GigaScience, 2022, 11. 4.Canaguier A, Guilbaud R, Denis E, et al. Oxford Nanopore and Bionano Genomics technologies evaluation for plant structural variation detection[J]. BMC genomics, 2022, 23(1): 1-17. 5.Shukla H, Suryamohan K, Khan A, et al. Near-chromosomal de novo assembly of Bengal tiger genome reveals genetic hallmarks of apex-predation[J]. bioRxiv, 2022. |
fingerprinting) 、脉冲场电泳(pulse field gel electrophoresis ,PFGE) 、限制性酶切分析( restriction ndonuclease fingerprinting ,REF) 、扩增片段长度多态性分析(am-lified fragment length polymororphism ,AFLP) 等进行链球菌的分型研究, 但这些技术存在技术复杂、耗时、分辨率低或重复性差等缺点。随机扩增多态性DNA 指纹(randomly am-lified polymorphic
 技术资料
技术资料暂无技术资料 索取技术资料