ChIP-Seq Data Analysis: Identification of ProteinDNA Binding Sites with SISSRs Peak-Finder
互联网
391
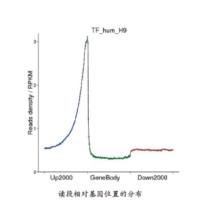
Protein–DNA interactions play key roles in determining gene-expression programs during cellular development and differentiation. Chromatin immunoprecipitation (ChIP) is the most widely used assay for probing such interactions. With recent advances in sequencing technology, ChIP-Seq, an approach that combines ChIP and next-generation parallel sequencing is fast becoming the method of choice for mapping protein–DNA interactions on a genome-wide scale. Here, we briefly review the ChIP-Seq approach for mapping protein–DNA interactions and describe the use of the SISSRs peak-finder, a software tool for precise identification of protein–DNA binding sites from sequencing data generated using ChIP-Seq.