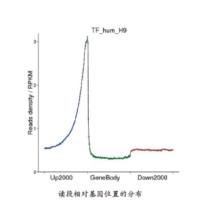
Epigenetic Analysis: ChIP-chip and ChIP-seq
互联网
1203
The access of transcription factors and the replication machinery to DNA is regulated by the epigenetic state of chromatin. In eukaryotes, this complex layer of regulatory processes includes the direct methylation of DNA, as well as covalent modifications to histones. Using next-generation sequencers, it is now possible to obtain profiles of epigenetic modifications across a genome using chromatin immunoprecipitation followed by sequencing (ChIP-seq). This technique permits the detection of the binding of proteins to specific regions of the genome with high resolution. It can be used to determine the target sequences of transcription factors, as well as the positions of histones with specific modification of their N-terminal tails. Antibodies that selectively bind methylated DNA may also be used to determine the position of methylated cytosines. Here, we present a data analysis pipeline for processing ChIP-seq data, and discuss the limitations and idiosyncrasies of these approaches.