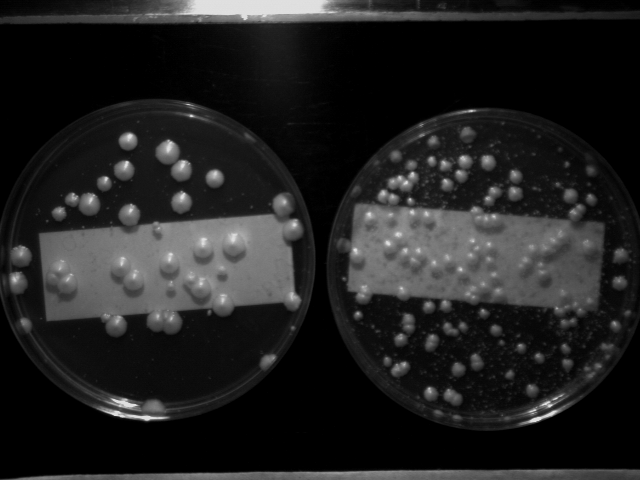
Typical transformation results
互联网
Typical transformation results
The following transformations were performed at the same time using the multiwell transformation protocol. Similar amounts of PCR product were used for each. The pictures demonstrate the degree of success and variability that can be attained. The plates were incubated for 6 days at 30°C before being photographed.
The haploid plate shows the many smaller background colonies (1 mm) which appear early during the incubation, in addition to the true transformants (3-10 mm). These smaller colonies will not show resistance to G418 when restreaked and do not appear in the diploid strain.
Colony size depends to some extent on the number of positives on the plate, on the amount of background and on whether the diprupted gene was important for viability.
The transformation efficiency of the diploid strain is approximately one-third of the haploid strain for unknown reasons. (One possiblity is that fewer diploid cells are used in the transformation per OD 600 than the haploid. This needs to be tested.)
DIPLOID-----------------------HAPLOID
Transformation results for ORF YER138C-A.
Gene name = TY2A
Gene product = TY2A protein
Previous disruption = unknown
The high transformation efficiency observed here is probably an artifact resulting from the fact that multiple TY elements exist in the genome. Similar results have been observed with certain telomeric open reading frames. The deletion primer picking algorithms have been updated to ensure that targeting primers are unique within the genome. If primers are not unique, then longer stretches of homologous sequence will be used in the primer design. This enhancement had not been implemented for the open reading frames which were disrupted in the trial stage.
DIPLOID-----------------------HAPLOID
Transformation results for ORF YER140W
Gene name = unassigned
Gene product = hypothetical protein
Previous disruption = unknown
DIPLOID-----------------------HAPLOID
Transformation results for ORF YER141W
Gene name = COX15
Gene product = cytochrome oxidase assembly factor
Previous disruption = unknown
This open reading frame might be a good candidate for an essential gene, based on transformation results. Recent data (Smith et al., Science 274 2069-2074), however, suggests that this orf may be disrupted, but results in a strain with a reduced growth rate. Therefore, it will be important to pick both big and small colonies for confirmation PCR.
DIPLOID-----------------------HAPLOID
Transformation results for ORF YER142C
Gene = MAG1
Gene product = 3-methyladenine DNA glycosylase
Previous disruption = viable