Mapping Genetic Polymorphisms Affecting Natural Variation in Drosophila Longevity
互联网
互联网
相关产品推荐

Recombinant-Schizosaccharomyces-pombe-Sphingosine-N-acyltransferase-lag1lag1Sphingosine N-acyltransferase lag1 EC= 2.3.1.24 Alternative name(s): Longevity assurance factor 1 Longevity assurance protein 1
¥12250

Recombinant-Mouse-Natural-killer-cells-antigen-CD94Klrd1Natural killer cells antigen CD94 Alternative name(s): Killer cell lectin-like receptor subfamily D member 1 CD_antigen= CD94
¥10374
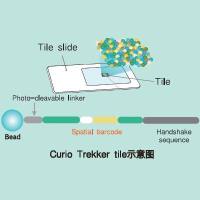
Trekker Single-Cell Spatial Mapping Kit 单细胞空间转录组分析试剂盒
询价

Recombinant-Saccharomyces-cerevisiae-Altered-inheritance-of-mitochondria-protein-11AIM11Altered inheritance of mitochondria protein 11 Alternative name(s): Genetic interactor of prohibitins 8
¥9996

Recombinant-Saccharomyces-cerevisiae-Sphingosine-N-acyltransferase-LAG1LAG1Sphingosine N-acyltransferase LAG1 EC= 2.3.1.24 Alternative name(s): Longevity assurance factor 1 Longevity assurance protein 1
¥12432
相关问答

