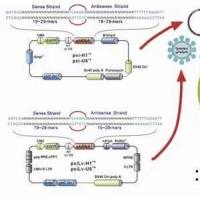
The pSico and pSicoR Design and Cloning
互联网
pSico Oligo Design
Click on the Mac OSX program pSicoOligomaker 1.5 to select and design oligos for pSico and plentilox3.7.
To operate it, simply paste your sequence in the "sequence" window, give it a name (optional), select a cutoff value (from -2 to 9) and click "search". The program will list in the right window all the potential 19-mers with a score equal or higher than the cutoff. The score is given based on a set of criteria published in Nature Biotech. by Reynolds et al. in 2004. According to the authors, a 19-mer with a value higher than 6 has approximately 90% chance of achieving silencing of the target mRNA.
You can select all targets and paste them in Word or Excel to store the result. In addition, if you type or paste a 19-mer in the field "target" and click on "make oligos" the program returns the sequence of the forward and reverse oligos (both in the 5' to 3' orientation) that have to be ordered to clone in pSico. The button "order" directs you to the web page we use to order oligos. The button "settings" lets you change the criteria to design the oligos (loop sequence and length, overhangs, etc) so that it can be used to design oligos for other vectors.
"Clear sequence" tells the program to remove all illicit characters, including spaces and "end of line", from the sequence. In addition, it converts the sequence to all caps.
In case you don't have a Mac, or you prefer to design your oligos manually, here is an example:
19mer: GTAGCTTAGCGTCGGAGCT
sense oligo:
5'-TGTAGCTTAGCGTCGGAGCT-TTCAAGAGA-AGCTCCGACGCTAAGCTACTTTTTTC
antisense oligo:
5'-TCGAGAAAAAAGTAGCTTAGCGTCGGAGCT-TCTCTTGAA-AGCTCCGACGCTAAGCTACA
How to clone into pSico and pSicoR:
We order 5’ phosphorylated, PAGE purified oligos designed using the program PSICOLIGOMAKER 1.5.
Oligos are resuspended in distilled water to a final concentration of 100µM.
Oligos annealing:
Mix:
-- ddH2O 23µl
-- sense oligo 1µl
-- antisense oligo 1µl
-- 2X Annealing Buffer 25µl (recipe see below)
• Incubate 4 minutes at 95æC
• Incubate 10 minutes at 70æC
• Slowly cool down the annealed oligos to 4æC (Store at �20æC)
Ligation:
• Dilute 1 µl of annealed oligos in 19 µl of water
• Ligate 1 µl of diluted annealed oligos to 50-100 ng of HpaI-XhoI digested pSico or pSicoRin a 10µl reaction. (Usually we do not dephosphorylate the vector, although it might help in case of partial digestion).
• Incubate at room temperature for 3 hours and transform 2µl of ligation.
• Digest minipreps with SacII-NotI (pSico) or XhoI-XbaI (pSicoR). Positive clones will release a fragment approximately 50 bp larger than the fragment released by the empty vector (710 vs 660 bp for pSico; 400 vs 350 bp for pSicoR). We use a 2% agarose gel to detect the shift. Positive clones should be sequence verified.
For pSico we use the following sequencing oligo (it is a reverse oligo and maps downstream to the cloning site):
5’- CAAACACAGTGCACACCACGC
For pSicoR we use the following sequencing oligo (it is a forward oligo and maps immediately upstream to the U6 promoter):
5’- TGCAGGGGAAAGAATAGTAGAC
2X Annealing Buffer:
200 mM potassium acetate
60 mM HEPES-KOH pH 7.4
4 mM Mg-acetate