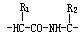A genetically encoded fluorescent amino acid(基因编码的荧光氨基酸)
互联网
Daniel Summerer
~undefinedDepartment of Chemistry, The Scripps Research Institute, 10550 North Torrey Pines Road, SR202, La Jolla, CA 92037; Beth Israel Deaconess Medical Center, Division of Signal Transduction, Harvard Medical School, 77 Avenue Louis Pasteur, Boston, MA 02115; Department of Chemistry, North Carolina State University, Campus Box 8240, Raleigh, NC 27695; and Division of Protein and Nucleic Acid Chemistry, Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, United Kingdom
molecular evolution | fluorescent probes | genetic code expansion | protein design | unnatural amino acids
Fluorescently labeled proteins are useful in a large number of bioanalytical applications, including in vivo imaging, high-throughput screening, diagnostics, proteomics, and single-biomolecule spectroscopy ( 1 – 5 ). Such applications have been greatly facilitated by the ability to label the protein of interest genetically with fluorescent proteins from jellyfish or coral ( 6 – 8 ). This method overcomes many of the difficulties associated with chemical modification, including yield, selectivity, and the need to modify proteins in vitro ( 9 ). However, the size of naturally occurring fluorescent proteins (>20 kDa) limits the resolution of distance measurements, and in many cases it represents a significant structural perturbation to the protein under study. Moreover, the fluorophore typically can be introduced only at the C or N terminus of a protein, and it is relatively insensitive to the local environment.
The use of small synthetic dyes allows investigation of local changes in distance or polarity with high precision and without significant structural perturbation of the protein. Nevertheless, most reactive dyes must be introduced into the protein in vitro , they exhibit relatively low chemoselectivity, and they are often limited to accessible positions at the protein surface ( 9 ). Moreover, mutagenesis of the target protein is often required to generate unique sites for derivatization. Recently, strategies have been developed to label proteins with small fluorescent dyes in vivo . One of these methods makes use of cell-permeant, biarsenic dyes that bind to their target protein through two arsenic–dithiol interactions ( 10 ). Another approach exploits a biotin ligase that recognizes a hydrazide-reactive ketone substrate that can be attached to a 15-mer acceptor peptide and subsequently linked to a hydrazide-derivatized fluorophore ( 11 ). It has also been shown that a highly promiscuous O 6 -alkylguanine-DNA-alkyltransferase, fused to a protein of interest, can be labeled by fluorescent O 6 -benzylguanosine derivatives ( 12 ). However, all of these approaches rely on the introduction of specific dye-acceptor motifs, peptides or proteins that restrict the sites of modification and can introduce undesired structural perturbations into the protein to be analyzed. Finally, in vitro mutagenesis with suppressor tRNAs chemically modified with fluorescent amino acids can be used to label proteins with fluorescent probes site-specifically, but this method is largely limited to in vitro systems affording small quantities of proteins ( 13 – 16 ).
Many of the challenges involved in the generation of fluorescently labeled proteins would be overcome if one could genetically encode fluorescent amino acids directly in prokaryotic or eukaryotic organisms. To this end, we report the efficient and selective biosynthetic introduction of a dansyl-containing amino acid into proteins in Saccharomyces cerevisiae in response to the amber nonsense codon, TAG.