Mapping Assembly Favored and Remodeled Nucleosome Positions on Polynucleosomal Templates
互联网
互联网
相关产品推荐

lptD/lptD蛋白Recombinant S_a_l_monella t_y_p_hi LPS-assembly protein LptD (lptD)重组蛋白lptD; imp; ostA; STY0108; t0096LPS-assembly protein LptD蛋白
¥2328
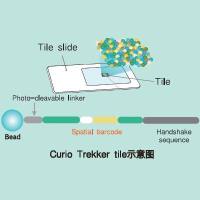
Trekker Single-Cell Spatial Mapping Kit 单细胞空间转录组分析试剂盒
询价

Recombinant-Vanderwaltozyma-polyspora-Spore-membrane-assembly-protein-2SMA2Spore membrane assembly protein 2
¥11998

Recombinant-Mouse-Mitofusin-2Mfn2Mitofusin-2 EC= 3.6.5.- Alternative name(s): Hypertension-related protein 1 Mitochondrial assembly regulatory factor; HSG protein Transmembrane GTPase MFN2
¥15512

Recombinant-Arabidopsis-thaliana-Surfeit-locus-protein-1-likeAt1g48510Surfeit locus protein 1-like; Surfeit 1-like Alternative name(s): Cytochrome c oxidase assembly protein SURF1-like
¥12194
相关问答

