25 In Organello Footprinting of mtDNA
互联网
903
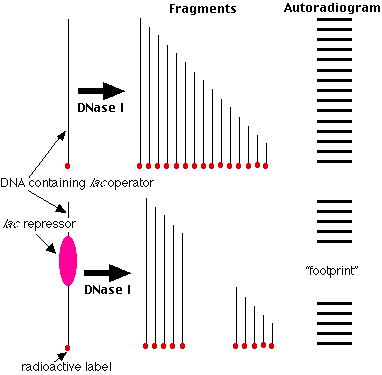
Studies of molecular mechanisms coordinating mammalian mitochondrial replicative and transcriptional processes have, for the most part, been limited to in vitro analyses. Although much has been learned from in vitro studies (1 ), they are often difficult to develop and may not depict biological processes occurring in vivo in a fully accurate manner. We have developed the method of mitochondrial DNA (mtDNA) footprinting as a means to analyze protein- mtDNA interactions within the isolated organelle, in an in vivo-like environment (2 –5 ). This procedure involves the use of dimethyl sulfate (DMS) in a methylation protection assay, similar to that used to analyze protein-nuclear DNA interactions in vivo (6 ). Dimethyl sulfate is a small molecule that methylates guanine residues at the N-7 position and, to a lesser extent, adenines at the N-3 position, making them sensitive to subsequent cleavage at alkaline pH and elevated temperature. When bound to specific DNA residues, proteins can decrease or intensify purine reactivity to DMS relative to naked DNA. The ability of DMS to readily permeate mitochondrial membranes permits detection of protein-mtDNA interactions that occur within purified organelles. We have employed two strategies (Southern hybridization and primer extension) to visualize and map alterations in mitochondrial DNA methylation resulting from protein binding in organello. Because the entire mitochondrial sequence of numerous species have been determined, it is possible to analyze virtually any mtDNA domain for protein interactions.