miRNA Data Analysis: Next-Gen Sequencing
互联网
775
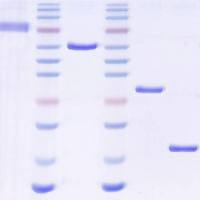
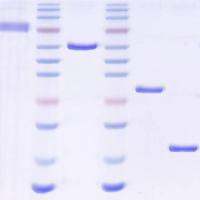
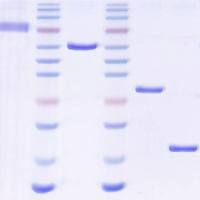
MicroRNAs (miRNAs) are short, noncoding RNAs that have the capacity to bind, capture, and silence hundreds of genes within and across diverse signaling pathways 1(Bartel, Cell 136:215–33, 2009) Specific sets of miRNAs characterize specific cell lineages of normal organisms and an increasing number of diseases have been shown to be associated with the dysregulation of specific miRNAs. Deep sequencing platforms have revealed unexpected complexity in relation to miRNAs, including 5′ and 3′-end-length heterogeneity and RNA editing. These insights not uncovered by previous microarray-based studies underscore the importance of data analysis tools that enable users to rapidly and easily analyze the unprecedented amounts of small RNA sequencing data that is emerging from next-generation sequencing platforms, such as Illumina/Solexa, SOLiD, and 454. In this chapter, we summarize the increasing number of analysis platforms that are available for miRNA discovery and profiling and the identification of functional miRNA–mRNA pairs in the context of biology and disease. We also discuss in greater detail our contributions to this effort.