DNA重组
CTAB TECHNIQUE / Method / Schedule / Protocol (JPB) FOR DNA ISOLATION / DNA EXTRACTION FROM PLANT LEAF / LEAVES SAMPLES (see also DNA RNA double isolation procedure if both DNA and RNA are needed) ...
CTAB TECHNIQUE / Method / Schedule / Protocol (JPB) FOR DNA ISOLATION / DNA EXTRACTION FROM PLANT LEAF / LEAVES SAMPLES (see also DNA RNA double isolation procedure if both DNA and RNA are needed) ...
grow plants in trays of 96 and leave two spots open (for the PCR controls) harvest 1 to 2 young and green leaves (1cm2/plant at rosette stage if possible). Use 96 well plates (1 or 2 ml E&K polypropyl ...
Overview This protocol describes a method for isolating DNA from plant tissue. Procedure1. Preheat the CTAB Isolation Buffer at 60°C. 2. Grind 2 g of fresh leaf tissue to a powder in Liquid Nitrogen ...
Overview This protocol describes a method for isolating DNA from plant tissue. Procedure1. Preheat the CTAB Isolation Buffer at 60°C. 2. Grind 2 g of fresh leaf tissue to a powder in Liquid Nitrogen ...
Streamlined DNA Extraction ProtocolThis method is derived from a procedure developed by Toby Bradshaw and the Poplar Molecular Genetics Cooperative. We have tested the procedure with a variety of Popu ...
ProcedureGrind in mortar and pestle or Waring blender with 5-7 volumes buffer A per g tissue. Use MCE at 350 l/L and if necessary with 5 ml 1 M DIECA/L. Squeeze through cheesecloth two layers of Mirac ...
Silica Clean-up of DNAMaterials:Silica Suspension: add 2 g of silica to 15 ml of H2Owash 3x by centifugation at 2000 x g for 2 minestimate vol of silica and resuspend in 2 vol H2O Silica Wash Solution ...
ProcedureGrind in mortar and pestle or Waring blender with 5-7 volumes buffer A per g tissue. Use MCE at 350 l/L and if necessary with 5 ml 1 M DIECA/L. Squeeze through cheesecloth two layers of Mirac ...
Phenol/Chloroform Precipitation of DNA1. Add an equal volume (equal to sample volume) of P/C to sample.2. Mix (shake don't vortex).3. Take aqueous (upper) layer. (If dirty sample repeat Ph/Chl step un ...
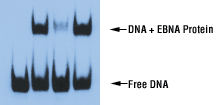
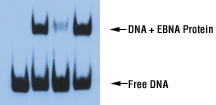
Gel Shift or Band Shift Assay or Electrophoretic Mobility Shift Assay (EMSA) is a technique for studying gene regulation and determining protein:DNA interactions. The assay is based on the observation ...
BAND-SHIFTThe procedure described in this chapter for the determination of affinity constants and kinetic dissociation constants by band-shift assay refers to an ideal antibody fragment (e.g. a scFv o ...
AbstractGene expression and regulation are mediated by DNA sequences in most instances directly upstream to the coding sequences by recruiting transcription factors regulators and a RNA polymerase in ...
EMSA using ds OligonucleotidesSolutions10X Annealing Buffer200 mM Tris 8.0 200 ml 1M Tris pH 8.010 mM EDTA 8.0 20 ml .5M EDTA pH 8.0500 mM NaCl 100 ml 5M NaCl280 ml Qstore at room temperature10X Kl ...
The interaction of proteins with DNA is central to the control of many cellular processes including DNA replication recombination and repair transcription and viral assembly. One technique that is cen ...
Gel Mobility Shift Assay for transcription factor bindingAuthor: Hueley LabSource: Contributed by Riddhish Shah Hueley LabAbstract: Protocol can be used to investigate binding of transcription factor ...
The interaction of proteins with DNA is central to the control of many cellular processes including DNA replication recombination and repair transcription and viral assembly. One technique that is cen ...
Gel Mobility Shift Assay Conditions -Mg/EDTA in Gel and BufferSteve Hahnlast modified Mon Nov 9 1998Protein Dilution Buffer5ml20 mM Tris pH7.9100 microliters 1M Tris 7.9150 mM KCl0.75 ml 1 M KCl1 mM D ...
Analysis of the interaction of proteins with either DNA or RNA sequences by in vivo footprinting involves two steps: (i) the in situ modification of nucleic acids by the footprinting reagent and (ii) ...
Overview An end labeled DNA probe is incubated with a purified DNA-binding factor or with a protein extract. The unprotected DNA is then digested with DNase I such that on average every DNA molecule i ...