Harvest (10000 rpm for 5 min) 24 h grown (in LB pH 7.5 37 oC 200 rpm) Escherichia coli Enterobacter aerogenes or Pseudomonas aeruginosa cells. Wash once with 0.05M potassium phosphate (KPi) bufferpH ...
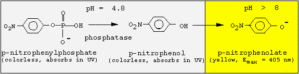
Phosphate esters are widely distributed in any organism. Nucleic acids metabolic intermediates like glucose-6-phosphate energy-rich substrates (AMP creatine phosphate) are some obvious examples. While ...
Typical modification sites: RGG box or RXR sequence motifs R-arginine G-glycineX-any aminoacid.Enzymes catalysing protein arginine methylation: PRMT1 from rat and the humanhomologue HRMT1L2 human PRMT ...
Purpose and backgroundsPrinciple of radioiodinationAddition of oxidizing reagents (such as chloramine-T or peroxidase+H202) converts I- to I+ or I3-. This highly reactive molecule attacks o-position o ...
Motor proteins of several kinesin family groups have now been crystallized: monomeric Kinesin-1 motor domains from human rat and Neurospora (Kull et al. 1996; Sack et al. 1997; Song et al. 2001) and d ...
Materials Milipore filter type HA 0.45 micronCulture plates (Linbro model 76-033-05)Protein in DDW or HEPES buffer (10 mg/mL)Vacuum grease Syringe with 18 G needleProcedure 1. Millipore filter all sol ...
Background:Proteins like many molecules can be prompted to form crystals when placed in the appropriate conditions. In order to crystallize a protein the purified protein undergoes slow precipitation ...
This specfic protocol is the latest incarnation of peptide mapping procedures that have been developed here in the TVL/MBVL of the Salk Institute over the last 20 years. Wade Gibson developed the firs ...
In identifying peptides from proteins with a known sequence it is often useful to be able to predict how a peptide will migrate during electrophoresis chromatography or two-dimensional peptide mapping ...
1. Immunoprecipitate the protein and run it on a preparative gel. CNBr cleavage must be done with protein transferred to a nitrocellulose filter. Neither Immobilon nor Nylon can substitute. IMPORTANT: ...
Materials: Overlay/Rehydration buffer:125 mM Tris-HCl pH 6.8 1mM EDTA 0.1% SDS 1mM 2-b-mercaptoethanol30% glycerolsome Bromophenol blueFor 1 ml you will need0.35 ml of 2X buffer0.30 ml H20 0.30 ml gly ...
1) Grow cells to a density of 5-8 X 105 cells/ml in RPMI 1640 containing serum.2) Pellet cells and wash 1 time with room temperature PBS.3) Resuspend final pellet in an appropriate volume of prewarmed ...
Buffers:- 2X buffer10 ml 0.5 M imidazol pH 6.60.21 g LiCl1.25 ml 1 M MgCl21.0 ml 0.1 M EGTA pH 6.6-- Bring volume up to 50 ml with distilled water.- dilution buffer1 ml 0.5 M imidazol pH 6.62.5 ml 20 ...
1) Remove 2 500 µl aliquots of supernatant into scintillation vials add scintillation fluid and count.2) Aliquot 1.6 mls of the remaining supernatant into each of two pyrex glass tubes.3) Add 6 mls of ...
Packing column:1) To 20 g of 100-200 mesh Bio-Sil A silica gel add 80 mls of chloroform.2) Place a small portion of glass wool at the base of the column.3) Pour gel solution into column and use a stir ...
1) Prepare a C-18 reverse phase prep-sep cartridge by washing with:a) 10 ml of methanolb) 20 ml of chloroform-methanol (2:1)c) 10 ml of methanold) 10 ml of methanol-1.6 M sodium acetate (1:1)2) Apply ...
Reagents:Bacterial strainE. coli N4830/pJW10LB amp media50 µg/ml ampicillinHigh salt bufferfor 1 L50 mM KH2PO4 6.8 g150 mM KCl 11.18 g50 mM sodium pyrophosphate (Na4P2O7-10H2O) 22.3 g5 mM Na2 EDTA 1.8 ...
1) Incubate cells with 1 µCi/ml of 3H-galactose for 72 hours.--- If treatment is for an extended period of time: treat in serum free media containing labeled galactose.2)Wash label out and spin cells ...
Reagents:Lysis buffer25 mM Tris-HCl pH 7.45 mM EDTA1 mM ATP20 µg/ml CLAP1 mM PMSFBuffer A10 mM MgCl20.2 M Tris-HCl pH 7.40.2 % Triton X-100Buffer B0.2 M sodium acetate pH 5.00.2 % Triton X-100Buffer C ...
As insects have an open circulatory system the hemolymph can be simply collected through an incision in the body wall. Most conveniently you should cut a leg and let the blood drip into a chilled glas ...