万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 保存条件:
-20℃
- 保质期:
6个月
- 英文名:
pIRES2-ZsGreen1
- 库存:
大量
- 供应商:
钰博生物
- 规格:
1ug/5ug
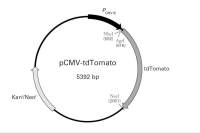
载体基本信息
| 出品公司: | Ybscience |
|---|---|
| 载体名称: | pIRES2-ZsGreen1 |
| 质粒类型: | 哺乳动物细胞载体;荧光报告载体;双顺反子载体 |
| 高拷贝/低拷贝: | 高拷贝 |
| 克隆方法: | 限制性内切酶,多克隆位点 |
| 启动子: | CMV |
| 载体大小: | 5283 bp |
| 5' 测序引物及序列: | CMV-F: 5'-CGCAAATGGGCGGTAGGCGTG-3' |
| 3' 测序引物及序列: | -- |
| 载体标签: | 绿色荧光蛋白ZsGreen1(C-端) |
| 载体抗性: | 卡那霉素 |
| 筛选标记: | 新霉素(Neomycin) |
| 克隆菌株: | DH5α, HB101 |
| 宿主细胞(系): | 常规细胞系,293、CV-1、CHO等 |
| 备注: | pIRES2-ZsGreen1载体含有IRES元件,目的基因与报告基因ZsGreen1共转录为一个mRNA,但分别翻译; ZsGreen1是亮度最高的绿色荧光蛋白,适合流式细胞仪筛选。 |
| 稳定性: | 瞬表达 或 稳表达 |
| 组成型/诱导型: | 组成型 |
| 病毒/非病毒: | 非病毒 |
载体质粒图谱和多克隆位点信息




载体简介
pIRES2-ZsGreen1载体描述 pIRES2-ZsGreen1 contains the internal ribosome entry site (IRES; 1, 2) of the encephalomyocarditis virus (ECMV) between an MCS and the Zoanthus sp. green fluorescent protein (ZsGreen; 3, 4)coding region. This design permits both the gene of interest (cloned into the MCS) and the ZsGreen1 gene to be translated from a single bicistronic mRNA. pIRES2-ZsGreen1 is designed for the efficient selection (by flow cytometry or other methods) of transiently transfected mammalian cells expressing ZsGreen1 and the protein of interest. This vector can be used to obtain stably transfected cell lines without time-consuming drug and clonal selection. ZsGreen1 is a human codon-optimized (5) ZsGreen variant that encodes the brightest commercially available green fluorescent protein. The MCS in pIRES2-ZsGreen1 is located between the immediate early promoter of cytomegalovirus (PCMV IE) and the IRES sequence. SV40 polyadenylation signals downstream of the ZsGreen1 gene direct proper processing of the 3' end of the bicistronic mRNA. The vector backbone also contains an SV40 origin for replication in mammalian cells expressing the SV40 T antigen. A neomycin-resistance cassette (Neor), consisting of the SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the herpes simplex virus thymidine kinase (HSV TK) gene, allows stably transfected eukaryotic cells to be selected using G418. A bacterial promoter upstream of this cassette expresses kanamycin resistance in E.coli. The pIRES2-ZsGreen1 backbone also provides a pUC origin of replication for propagation in E. coli and an f1 origin for single-stranded DNA production. pIRES2-ZsGreen1 can be used to quickly identify cells expressing a gene of interest by screening for ZsGreen1 fluorescence. Genes inserted into the MCS should include an initiating ATG codon. Selection of ZsGreen1-positive cells is possible 24–36 hours after transfection by flow cytometry or fluorescence microscopy. pIRES2-ZsGreen1 and its derivatives can be introduced into mammalian cells using any standard transfection method. If required, stable transformants can be selected using G418 (6). Selection of Stable Transfectants Selectable marker: plasmid confers resistance to G418 (500 μg/ml) Propagation in E. coli Suitable host strains: DH5α, HB101, and other general purpose strains. Single-stranded DNA production requires a host containing an F plasmid such as JM101 or XL1-Blue. Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts. E. coli replication origin: pUC Copy number: high
载体序列
LOCUS pIRES2-ZsGreen1 5283 bp DNA SYN DEFINITION pIRES2-ZsGreen1 ACCESSION KEYWORDS SOURCE ORGANISM other sequences; artificial sequences; vectors. FEATURES Location/Qualifiers source 1..5283 /organism="pIRES2-ZsGreen1" /mol_type="other DNA" promoter 10..562 /label="CMV_immearly_promoter" misc_feature 65..352 /label="CAG_enhancer" misc_feature 519..539 /label="CMV_fwd_primer" promoter 520..589 /label="CMV_promoter" misc_feature 666..1250 /label="IRES" CDS 1242..1949 /label="ORF frame 3" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" gene 1257..1946 /label="Zs_Green" /gene="Zs_Green" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" misc_feature 2162..2181 /label="EBV_rev_primer" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" rep_origin complement(2331..2637) /label="f1_origin" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" promoter 2716..2744 /label="AmpR_promoter" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" misc_feature complement(2810..2830) /label="pBABE_3_primer" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" misc_feature complement(2816..3031) /label="SV40_enhancer" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" promoter 2828..3096 /label="SV40_promoter" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" rep_origin 2995..3072 /label="SV40_origin" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" misc_feature 3057..3076 /label="SV40pro_F_primer" /translation="MATTMAQSKHGLTKEMTMKYRMEGCVDGHKFVITGEGIGYPFKG KQAINLCVVEGGPLPFAEDILSAAFMYGNRVFTEYPQDIVDYFKNSCPAGYTWDRSFL FEDGAVCICNADITVSVEENCMYHESKFYGVNFPADGPVMKKMTDNWEPSCEKIIPVP KQGILKGDVSMYLLLKDGGRLRCQFDTVYKAKSVPRKMPDWHFIQHKLTREDRSDAKN QKWHLTEHAIASGSALP*" CDS 3179..3973 /label="ORF frame 2" /translation="MIEQDGLHAGSPAAWVERLFGYDWAQQTIGCSDAAVFRLSAQGR PVLFVKTDLSGALNELQDEAARLSWLATTGVPCAAVLDVVTEAGRDWLLLGEVPGQDL LSSHLAPAEKVSIMADAMRRLHTLDPATCPFDHQAKHRIERARTRMEAGLVDQDDLDE EHQGLAPAELFARLKASMPDGEDLVVTHGDACLPNIMVENGRFSGFIDCGRLGVADRY QDIALATRDIAEELGGEWADRFLVLYGIAAPDSQRIAFYRLLDEFF*" gene 3182..3970 /label="NeoR/KanR" /gene="NeoR/KanR" /translation="MIEQDGLHAGSPAAWVERLFGYDWAQQTIGCSDAAVFRLSAQGR PVLFVKTDLSGALNELQDEAARLSWLATTGVPCAAVLDVVTEAGRDWLLLGEVPGQDL LSSHLAPAEKVSIMADAMRRLHTLDPATCPFDHQAKHRIERARTRMEAGLVDQDDLDE EHQGLAPAELFARLKASMPDGEDLVVTHGDACLPNIMVENGRFSGFIDCGRLGVADRY QDIALATRDIAEELGGEWADRFLVLYGIAAPDSQRIAFYRLLDEFF*" CDS complement(3488..4024) /label="ORF frame 3" /translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ PPHCISHDGYFLGRSKVR*" terminator 4148..4417 /label="TK_PA_terminator" /translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ PPHCISHDGYFLGRSKVR*" rep_origin 4565..5184 /label="pBR322_origin" /translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ PPHCISHDGYFLGRSKVR*" ORIGIN 1 TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA TGGAGTTCCG 61 CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC CCCGCCCATT 121 GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC ATTGACGTCA 181 ATGGGTGGAG TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT ATCATATGCC 241 AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT ATGCCCAGTA 301 CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA TCGCTATTAC 361 CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG ACTCACGGGG 421 ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC AAAATCAACG 481 GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG GTAGGCGTGT 541 ACGGTGGGAG GTCTATATAA GCAGAGCTGG TTTAGTGAAC CGTCAGATCC GCTAGCGCTA 601 CCGGACTCAG ATCTCGAGCT CAAGCTTCGA ATTCTGCAGT CGACGGTACC GCGGGCCCGG 661 GATCCGCCCC TCTCCCTCCC CCCCCCCTAA CGTTACTGGC CGAAGCCGCT TGGAATAAGG 721 CCGGTGTGCG TTTGTCTATA TGTTATTTTC CACCATATTG CCGTCTTTTG GCAATGTGAG 781 GGCCCGGAAA CCTGGCCCTG TCTTCTTGAC GAGCATTCCT AGGGGTCTTT CCCCTCTCGC 841 CAAAGGAATG CAAGGTCTGT TGAATGTCGT GAAGGAAGCA GTTCCTCTGG AAGCTTCTTG 901 AAGACAAACA ACGTCTGTAG CGACCCTTTG CAGGCAGCGG AACCCCCCAC CTGGCGACAG 961 GTGCCTCTGC GGCCAAAAGC CACGTGTATA AGATACACCT GCAAAGGCGG CACAACCCCA 1021 GTGCCACGTT GTGAGTTGGA TAGTTGTGGA AAGAGTCAAA TGGCTCTCCT CAAGCGTATT 1081 CAACAAGGGG CTGAAGGATG CCCAGAAGGT ACCCCATTGT ATGGGATCTG ATCTGGGGCC 1141 TCGGTACACA TGCTTTACAT GTGTTTAGTC GAGGTTAAAA AAACGTCTAG GCCCCCCGAA 1201 CCACGGGGAC GTGGTTTTCC TTTGAAAAAC ACGATGATAA TATGGCCACA ACCATGGCCC 1261 AGTCCAAGCA CGGCCTGACC AAGGAGATGA CCATGAAGTA CCGCATGGAG GGCTGCGTGG 1321 ACGGCCACAA GTTCGTGATC ACCGGCGAGG GCATCGGCTA CCCCTTCAAG GGCAAGCAGG 1381 CCATCAACCT GTGCGTGGTG GAGGGCGGCC CCTTGCCCTT CGCCGAGGAC ATCTTGTCCG 1441 CCGCCTTCAT GTACGGCAAC CGCGTGTTCA CCGAGTACCC CCAGGACATC GTCGACTACT 1501 TCAAGAACTC CTGCCCCGCC GGCTACACCT GGGACCGCTC CTTCCTGTTC GAGGACGGCG 1561 CCGTGTGCAT CTGCAACGCC GACATCACCG TGAGCGTGGA GGAGAACTGC ATGTACCACG 1621 AGTCCAAGTT CTACGGCGTG AACTTCCCCG CCGACGGCCC CGTGATGAAG AAGATGACCG 1681 ACAACTGGGA GCCCTCCTGC GAGAAGATCA TCCCCGTGCC CAAGCAGGGC ATCTTGAAGG 1741 GCGACGTGAG CATGTACCTG CTGCTGAAGG ACGGTGGCCG CTTGCGCTGC CAGTTCGACA 1801 CCGTGTACAA GGCCAAGTCC GTGCCCCGCA AGATGCCCGA CTGGCACTTC ATCCAGCACA 1861 AGCTGACCCG CGAGGACCGC AGCGACGCCA AGAACCAGAA GTGGCACCTG ACCGAGCACG 1921 CCATCGCCTC CGGCTCCGCC TTGCCCTGAG CGGCCGCGAC TCTAGATCAT AATCAGCCAT 1981 ACCACATTTG TAGAGGTTTT ACTTGCTTTA AAAAACCTCC CACACCTCCC CCTGAACCTG 2041 AAACATAAAA TGAATGCAAT TGTTGTTGTT AACTTGTTTA TTGCAGCTTA TAATGGTTAC 2101 AAATAAAGCA ATAGCATCAC AAATTTCACA AATAAAGCAT TTTTTTCACT GCATTCTAGT 2161 TGTGGTTTGT CCAAACTCAT CAATGTATCT TAAGGCGTAA ATTGTAAGCG TTAATATTTT 2221 GTTAAAATTC GCGTTAAATT TTTGTTAAAT CAGCTCATTT TTTAACCAAT AGGCCGAAAT 2281 CGGCAAAATC CCTTATAAAT CAAAAGAATA GACCGAGATA GGGTTGAGTG TTGTTCCAGT 2341 TTGGAACAAG AGTCCACTAT TAAAGAACGT GGACTCCAAC GTCAAAGGGC GAAAAACCGT 2401 CTATCAGGGC GATGGCCCAC TACGTGAACC ATCACCCTAA TCAAGTTTTT TGGGGTCGAG 2461 GTGCCGTAAA GCACTAAATC GGAACCCTAA AGGGAGCCCC CGATTTAGAG CTTGACGGGG 2521 AAAGCCGGCG AACGTGGCGA GAAAGGAAGG GAAGAAAGCG AAAGGAGCGG GCGCTAGGGC 2581 GCTGGCAAGT GTAGCGGTCA CGCTGCGCGT AACCACCACA CCCGCCGCGC TTAATGCGCC 2641 GCTACAGGGC GCGTCAGGTG GCACTTTTCG GGGAAATGTG CGCGGAACCC CTATTTGTTT 2701 ATTTTTCTAA ATACATTCAA ATATGTATCC GCTCATGAGA CAATAACCCT GATAAATGCT 2761 TCAATAATAT TGAAAAAGGA AGAGTCCTGA GGCGGAAAGA ACCAGCTGTG GAATGTGTGT 2821 CAGTTAGGGT GTGGAAAGTC CCCAGGCTCC CCAGCAGGCA GAAGTATGCA AAGCATGCAT 2881 CTCAATTAGT CAGCAACCAG GTGTGGAAAG TCCCCAGGCT CCCCAGCAGG CAGAAGTATG 2941 CAAAGCATGC ATCTCAATTA GTCAGCAACC ATAGTCCCGC CCCTAACTCC GCCCATCCCG 3001 CCCCTAACTC CGCCCAGTTC CGCCCATTCT CCGCCCCATG GCTGACTAAT TTTTTTTATT 3061 TATGCAGAGG CCGAGGCCGC CTCGGCCTCT GAGCTATTCC AGAAGTAGTG AGGAGGCTTT 3121 TTTGGAGGCC TAGGCTTTTG CAAAGATCGA TCAAGAGACA GGATGAGGAT CGTTTCGCAT 3181 GATTGAACAA GATGGATTGC ACGCAGGTTC TCCGGCCGCT TGGGTGGAGA GGCTATTCGG 3241 CTATGACTGG GCACAACAGA CAATCGGCTG CTCTGATGCC GCCGTGTTCC GGCTGTCAGC 3301 GCAGGGGCGC CCGGTTCTTT TTGTCAAGAC CGACCTGTCC GGTGCCCTGA ATGAACTGCA 3361 AGACGAGGCA GCGCGGCTAT CGTGGCTGGC CACGACGGGC GTTCCTTGCG CAGCTGTGCT 3421 CGACGTTGTC ACTGAAGCGG GAAGGGACTG GCTGCTATTG GGCGAAGTGC CGGGGCAGGA 3481 TCTCCTGTCA TCTCACCTTG CTCCTGCCGA GAAAGTATCC ATCATGGCTG ATGCAATGCG 3541 GCGGCTGCAT ACGCTTGATC CGGCTACCTG CCCATTCGAC CACCAAGCGA AACATCGCAT 3601 CGAGCGAGCA CGTACTCGGA TGGAAGCCGG TCTTGTCGAT CAGGATGATC TGGACGAAGA 3661 GCATCAGGGG CTCGCGCCAG CCGAACTGTT CGCCAGGCTC AAGGCGAGCA TGCCCGACGG 3721 CGAGGATCTC GTCGTGACCC ATGGCGATGC CTGCTTGCCG AATATCATGG TGGAAAATGG 3781 CCGCTTTTCT GGATTCATCG ACTGTGGCCG GCTGGGTGTG GCGGACCGCT ATCAGGACAT 3841 AGCGTTGGCT ACCCGTGATA TTGCTGAAGA GCTTGGCGGC GAATGGGCTG ACCGCTTCCT 3901 CGTGCTTTAC GGTATCGCCG CTCCCGATTC GCAGCGCATC GCCTTCTATC GCCTTCTTGA 3961 CGAGTTCTTC TGAGCGGGAC TCTGGGGTTC GAAATGACCG ACCAAGCGAC GCCCAACCTG 4021 CCATCACGAG ATTTCGATTC CACCGCCGCC TTCTATGAAA GGTTGGGCTT CGGAATCGTT 4081 TTCCGGGACG CCGGCTGGAT GATCCTCCAG CGCGGGGATC TCATGCTGGA GTTCTTCGCC 4141 CACCCTAGGG GGAGGCTAAC TGAAACACGG AAGGAGACAA TACCGGAAGG AACCCGCGCT 4201 ATGACGGCAA TAAAAAGACA GAATAAAACG CACGGTGTTG GGTCGTTTGT TCATAAACGC 4261 GGGGTTCGGT CCCAGGGCTG GCACTCTGTC GATACCCCAC CGAGACCCCA TTGGGGCCAA 4321 TACGCCCGCG TTTCTTCCTT TTCCCCACCC CACCCCCCAA GTTCGGGTGA AGGCCCAGGG 4381 CTCGCAGCCA ACGTCGGGGC GGCAGGCCCT GCCATAGCCT CAGGTTACTC ATATATACTT 4441 TAGATTGATT TAAAACTTCA TTTTTAATTT AAAAGGATCT AGGTGAAGAT CCTTTTTGAT 4501 AATCTCATGA CCAAAATCCC TTAACGTGAG TTTTCGTTCC ACTGAGCGTC AGACCCCGTA 4561 GAAAAGATCA AAGGATCTTC TTGAGATCCT TTTTTTCTGC GCGTAATCTG CTGCTTGCAA 4621 ACAAAAAAAC CACCGCTACC AGCGGTGGTT TGTTTGCCGG ATCAAGAGCT ACCAACTCTT 4681 TTTCCGAAGG TAACTGGCTT CAGCAGAGCG CAGATACCAA ATACTGTTCT TCTAGTGTAG 4741 CCGTAGTTAG GCCACCACTT CAAGAACTCT GTAGCACCGC CTACATACCT CGCTCTGCTA 4801 ATCCTGTTAC CAGTGGCTGC TGCCAGTGGC GATAAGTCGT GTCTTACCGG GTTGGACTCA 4861 AGACGATAGT TACCGGATAA GGCGCAGCGG TCGGGCTGAA CGGGGGGTTC GTGCACACAG 4921 CCCAGCTTGG AGCGAACGAC CTACACCGAA CTGAGATACC TACAGCGTGA GCTATGAGAA 4981 AGCGCCACGC TTCCCGAAGG GAGAAAGGCG GACAGGTATC CGGTAAGCGG CAGGGTCGGA 5041 ACAGGAGAGC GCACGAGGGA GCTTCCAGGG GGAAACGCCT GGTATCTTTA TAGTCCTGTC 5101 GGGTTTCGCC ACCTCTGACT TGAGCGTCGA TTTTTGTGAT GCTCGTCAGG GGGGCGGAGC 5161 CTATGGAAAA ACGCCAGCAA CGCGGCCTTT TTACGGTTCC TGGCCTTTTG CTGGCCTTTT 5221 GCTCACATGT TCTTTCCTGC GTTATCCCCT GATTCTGTGG ATAACCGTAT TACCGCCATG 5281 CAT
风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验相关专题 总有一种转染方法适合你 参与者:水仙子2005 最近用pIRES2-eGFP做转染 ,可是看不到荧光(就连空载体也没有),由于实验要在3月底出结果,所以现在很急,准备寒假加班加点做。 有人说这个载体 的GFP表达很不稳定,不知道有没有人用过这个载体,能否给点建议,对于好的建议愿以积分相赠。 参与者:Crazyvirus 不知你是固定以后观察荧光还是转染 后直接看的,若是固定
pSUPEREGFP表达 绿色 荧光蛋白和目的基因的融合蛋白,目的基因位于N端 kan/neo 4.7kb E. coli/mammals pIRES2-EGFP pIRES2EGFP 双顺反子表达目的基因和 绿色 荧光蛋白 kan/neo 5.3kb E. coli/mammals pIRES2-ZsGreen1 双顺反子表达目的基因和 绿色 荧光蛋白,目的基因位于C端 kan/neo 5.3kb E. coli/mammals pIRES-hrGFP-1a pDsred1-N1
sunyuzhu10 包装质粒是pMD2.G pMDLg/pRRE pRSV-Rev 过表达载体是pLVX-IRES-ZsGreen1 这样的四个质粒能包装出慢病毒么? 请有经验的战友帮忙看下 暴君 可以的 644414915 pMD2.G是VSVG吗 我现在正在包病毒,别人给了我这四个质粒,别的什么都没说,我不知道该怎么包装装染 谁有具体步骤 转染
 技术资料
技术资料暂无技术资料 索取技术资料