相关产品推荐更多 >
万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 英文名:
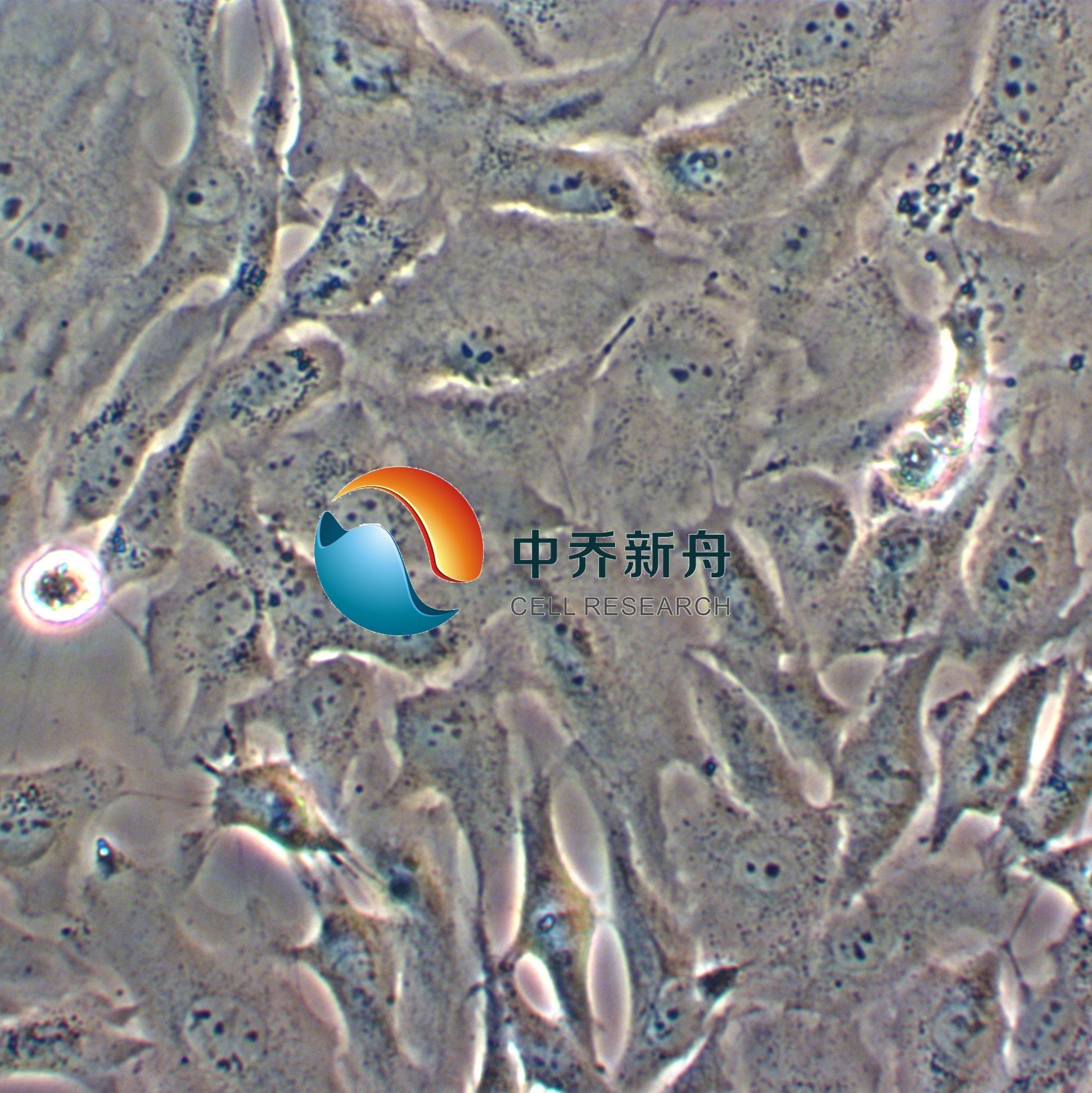
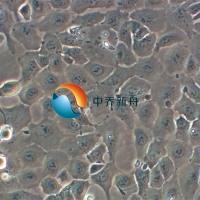
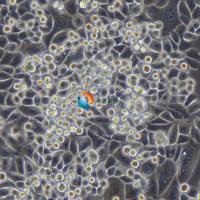
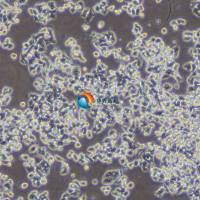
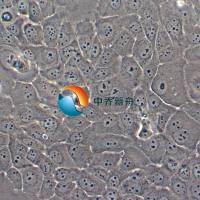
Calu-1
- 库存:
大量
- 供应商:
中乔新舟
- 细胞类型:
细胞系
- 品系:
human
- 组织来源:
human
- 相关疾病:
否
- 物种来源:
human
- 免疫类型:
否
- 细胞形态:
咨询销售
- 器官来源:
human
- 运输方式:
T25瓶运输
- 年限:
5-10年
- 生长状态:
贴壁生长
- 规格:
5 x 10^5 cells/vial
|
产品名称 |
Calu-1人肺癌细胞 |
|
货号 |
ZQ0394 |
|
产品介绍 |
Calu-1 是一种表现出上皮形态的细胞系,从一名 47 岁的表皮样癌白人男性患者的肺部分离出来,其超微结构特征包括众多微绒毛,显著的RER(粗面内质网),溶酶体,脂包含体,无病毒颗粒, 含ras (H-ras)癌基因,已广泛应用于肺癌生物学、药物筛选和细胞毒性研究。Calu-1细胞表达肺上皮细胞特有的几种标志物,已成为研究肺癌发生和治疗耐药分子通路的有价值的模型。 |
|
种属 |
人 |
|
性别/年龄 |
男 |
|
组织 |
肺;转移灶:肋膜 |
|
疾病 |
III期,表皮瘤 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮的 |
|
生长方式 |
贴壁 |
|
倍增时间 |
大约37~49.8小时 |
|
培养基和添加剂 |
McCOY's 5A(品牌:中乔新舟 货号:ZQ-1000)+10%胎牛血清(中乔新舟 货号:ZQ0500)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
ZM0394 |
|
生物安全等级 |
BSL-1 |
|
STR位点信息 |
Amelogenin: X |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; HTB-54 ECACC; 93120818 |
|
供应限制 |
仅供科研使用 |
上海中乔新舟生物科技有限公司成立于2011年,历经十多年发展,主要专注于细胞生物学产品的研究和开发,专注于为药企、各类科研机构及CRO企业提供符合标准规范的细胞培养服务、细胞培养基、细胞检测试剂盒、细胞培养试剂,胎牛血清和细胞生物学技术服务等。
公司一直致力于为高等院校、研究机构、医院、CRO及CDMO企业提供细胞培养完整解决方案,这些产品旨在满足细胞培养的多样需求,确保实验和研究的有效进行。引用中乔新舟(ZQXZBIO)产品和服务的文献超数千篇。

产品服务
细胞资源:原代细胞、细胞株、干细胞、示踪细胞、耐药株细胞、永生化细胞等基因工程细胞。
试剂产品:胎牛血清、完全培养基(适用于原代细胞及细胞株)、无血清培养基、基础培养基、细胞转染试剂、重组因子、胰酶和双抗等等细胞培养所有实验相关产品。
技术服务:稳转株构建、原代细胞分离、特殊培养基定制服务、细胞检测等。

目前产品已经畅销国内30多个省市,与客户建立长期的合作伙伴关系,共同实现成功。全体员工将不懈努力,继续为科研人员提供优良的产品和服务,致力成为全球细胞培养领域的参与者。
企业愿景
致力于成为国内细胞培养基产业的佼佼者,生物医药领域上游原材料的优良提供商。
企业使命
成长为专业细胞系及原代细胞培养供应商、专业细胞培养基及培养试剂生产商。
企业荣誉


风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验DOI=10.1007/978-1-4757-1647-4_5
Fogh J., Trempe G.L.
New human tumor cell lines.
(In) Human tumor cells in vitro; Fogh J. (eds.); pp.115-159; Springer; New York (1975)
PubMed=327080; DOI=10.1093/jnci/59.1.221
Fogh J., Fogh J.M., Orfeo T.
One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice.
J. Natl. Cancer Inst. 59:221-226(1977)
PubMed=833871; DOI=10.1093/jnci/58.2.209
Fogh J., Wright W.C., Loveless J.D.
Absence of HeLa cell contamination in 169 cell lines derived from human tumors.
J. Natl. Cancer Inst. 58:209-214(1977)
PubMed=6935474; DOI=10.1093/jnci/66.2.239
Wright W.C., Daniels W.P., Fogh J.
Distinction of seventy-one cultured human tumor cell lines by polymorphic enzyme analysis.
J. Natl. Cancer Inst. 66:239-247(1981)
PubMed=7017212; DOI=10.1093/jnci/66.6.1003
Pollack M.S., Heagney S.D., Livingston P.O., Fogh J.
HLA-A, B, C and DR alloantigen expression on forty-six cultured human tumor cell lines.
J. Natl. Cancer Inst. 66:1003-1012(1981)
PubMed=7459858
Rousset M., Zweibaum A., Fogh J.
Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins.
Cancer Res. 41:1165-1170(1981)
PubMed=6582512; DOI=10.1073/pnas.81.2.568
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=3518877; DOI=10.3109/07357908609038260
Fogh J.
Human tumor lines for cancer research.
Cancer Invest. 4:157-184(1986)
PubMed=8385084; DOI=10.1111/j.1349-7006.1993.tb02851.x
Suzuki S., Takahashi T., Nakamura S., Koike K., Ariyoshi Y., Takahashi T., Ueda R.
Alterations of integrin expression in human lung cancer.
Jpn. J. Cancer Res. 84:168-174(1993)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=7736387; DOI=10.1002/1097-0142(19950515)75:10<2442::AID-CNCR2820751009>3.0.CO;2-Q
Campling B.G., Sarda I.R., Baer K.A., Pang S.C., Baker H.M., Lofters W.S., Flynn T.G.
Secretion of atrial natriuretic peptide and vasopressin by small cell lung cancer.
Cancer 75:2442-2451(1995)
PubMed=11583962; DOI=10.1016/S0002-9440(10)62521-7
Haruki N., Harano T., Masuda A., Kiyono T., Takahashi T., Tatematsu Y., Shimizu S., Mitsudomi T., Konishi H., Osada H., Fujii Y., Takahashi T.
Persistent increase in chromosome instability in lung cancer: possible indirect involvement of p53 inactivation.
Am. J. Pathol. 159:1345-1352(2001)
PubMed=12068308; DOI=10.1038/nature00766
Davies H., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=12794755; DOI=10.1002/ijc.11184
Endoh H., Yatabe Y., Shimizu S., Tajima K., Kuwano H., Takahashi T., Mitsudomi T.
RASSF1A gene inactivation in non-small cell lung cancer and its clinical implication.
Int. J. Cancer 106:45-51(2003)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22961666; DOI=10.1158/2159-8290.CD-12-0112
Byers L.A., Wang J., Nilsson M.B., Fujimoto J., Saintigny P., Yordy J., Giri U., Peyton M., Fan Y.-H., Diao L.-X., Masrorpour F., Shen L., Liu W.-B., Duchemann B., Tumula P., Bhardwaj V., Welsh J., Weber S., Glisson B.S., Kalhor N., Wistuba I.I., Girard L., Lippman S.M., Mills G.B., Coombes K.R., Weinstein J.N., Minna J.D., Heymach J.V.
Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1.
Cancer Discov. 2:798-811(2012)
PubMed=24002593; DOI=10.1038/bjc.2013.452
Wu D., Pang Y., Wilkerson M.D., Wang D., Hammerman P.S., Liu J.S.
Gene-expression data integration to squamous cell lung cancer subtypes reveals drug sensitivity.
Br. J. Cancer 109:1599-1608(2013)
PubMed=25984343; DOI=10.1038/sdata.2014.35
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
littleyan0418 我现在的任务是要构建人肺癌细胞的cDNA文库,但我不是这个方面科班出身,好多地方还不是很明白,想请教师兄师姐的经验,应该注意些什么。现在对总RNA的提取我有点找不出头绪。麻烦师兄师姐们可以给我分享些这方面的资料和网址吗?非常感谢!! neuronboy http://www.takara.com.cn/ 价格、试剂盒和说明书都有。呵呵 littleyan0418
Transepithelial Measurements of Bicarbonate Secretion in Calu-3 Cells
poorly understood and underinvestigated. Studies from our own laboratory ( 4 ) and the laboratories of Wine and Widdicombe ( 5 ) have now established that the human airway serous cell line, Calu-3 cells, secrete HCO 3 and not Cl - , in response to a cAMP
我培养过不少肺癌细胞(人类),其中绝大部分都是贴壁细胞,具体如: (1) A549(肺腺癌) (2) NCIH446(小肺细胞癌) (3) 801(非小肺细胞癌) (4) NCIH460 (大细胞肺癌) 在消化传代过程中,步骤基本一致: 吸去旧的培养液->用Hanks'(1X)清洗一两次->加入一定量的消化液(Trpsin-EDTA)->置显微镜下观察,待细胞大部分变圆时,回到超静台->吸去消化液->加入一定量的新培养液->反复吹打细胞->再置显微镜下观察,直到细胞全部悬浮起来->
 技术资料
技术资料暂无技术资料 索取技术资料