| 细胞名称: | Hs 746.T细胞, ATCC HTB-135 细胞, Hs746T细胞, 人胃癌细胞 |
|---|---|
| 细胞又名: | Hs 746T; HS 746T; Hs-746T; HS-746T; Hs746T; HS746T; Hs746-T; 746T |
| 细胞来源: | ATCC |
| 产品货号: | HTB-135 |
| 种属来源: | 人 |
| 组织来源: | 胃 |
| 患者年龄: | 74 |
| 患者性别: | 男 |
| 细胞起源: | Hs 746.T于1971年从一个56岁的白人男性患者的左锁骨上区转移性肿瘤结节的分离获得,经组织学证实诊断为结肠腺癌。 |
| 抗原表达: | HLA A11, B15, B17, Cw1, Cw3; blood type B |
| 癌基因: | myc +; myb + ; ras +; fos +; p53 +; sis -; abl -; ros -; src - |
| 基因表达: | 癌胚抗原(CEA)908 ng/106个细胞/10天。CSAp(CSAp-)和结肠抗原3表达为阴性。 |
| 癌细胞诱导: | 能够诱导癌细胞产生 |
| 诱导实验: |
是的,在免疫抑制的老鼠身上
(裸鼠皮下接种10的7次方个细胞后,21天内肿瘤以100%的形成(5/5))
|
| 细胞形态: | 上皮样细胞 |
| 生长特性: | 贴壁生长 |
| 培养基: | DMEM培养基,90%;FBS,10%。 |
| 存储人: | M Romsdahl |
| 生长条件: | 气相:空气,95%;二氧化碳,5%; 温度:37 ℃, |
| 传代方法: | 1:2至1:8,每周2次。 |
| 冻存条件: | 95% 完全培养基+5% DMSO,液氮储存 |
| 支原体检测: | 阴性 |
| 安全等级: | 1 |
| 应用: | Hs 746T在细胞的大小、形状和染色上都有明显的改变,细胞核和核仁形状都很奇怪。克隆细胞能够在正常人上皮细胞和成纤维细胞单层细胞上以及纤维素膜上聚集。 |
| STR: |
Amelogenin: X,Y
CSF1PO: 10,12
D13S317: 10
D16S539: 11
D5S818: 11,13
D7S820: 8
TH01: 9
TPOX: 8,10
vWA: 15,17
|
| 细胞说明: | 在不同的早期传代能检测到体外转化特性。Hs 746T在细胞的大小、形状和染色上都有明显的改变,细胞核和核仁形状都很奇怪。克隆细胞能够在正常人上皮细胞和成纤维细胞单层细胞上以及纤维素膜上聚集。 |
| 参考文献: |
1.Dutil J., Chen Z., Monteiro A.N., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
2.Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X., Egan R.K., Liu Q., Miroo T., Mitropoulos X., Richardson L., Wang J., Zhang T., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
3.Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
4.Liu J., McCleland M., Stawiski E.W., Gnad F., Mayba O., Haverty P.M., Durinck S., Chen Y.-J., Klijn C., Jhunjhunwala S., Lawrence M., Liu H., Wan Y., Chopra V., Yaylaoglu M.B., Yuan W., Ha C., Gilbert H.N., Reeder J., Pau G., Stinson J., Stern H.M., Manning G., Wu T.D., Neve R.M., de Sauvage F.J., Modrusan Z., Seshagiri S., Firestein R., Zhang Z.
Integrated exome and transcriptome sequencing reveals ZAK isoform usage in gastric cancer.
Nat. Commun. 5:3830-3830(2014)
5.Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
|
| 细胞图片: |
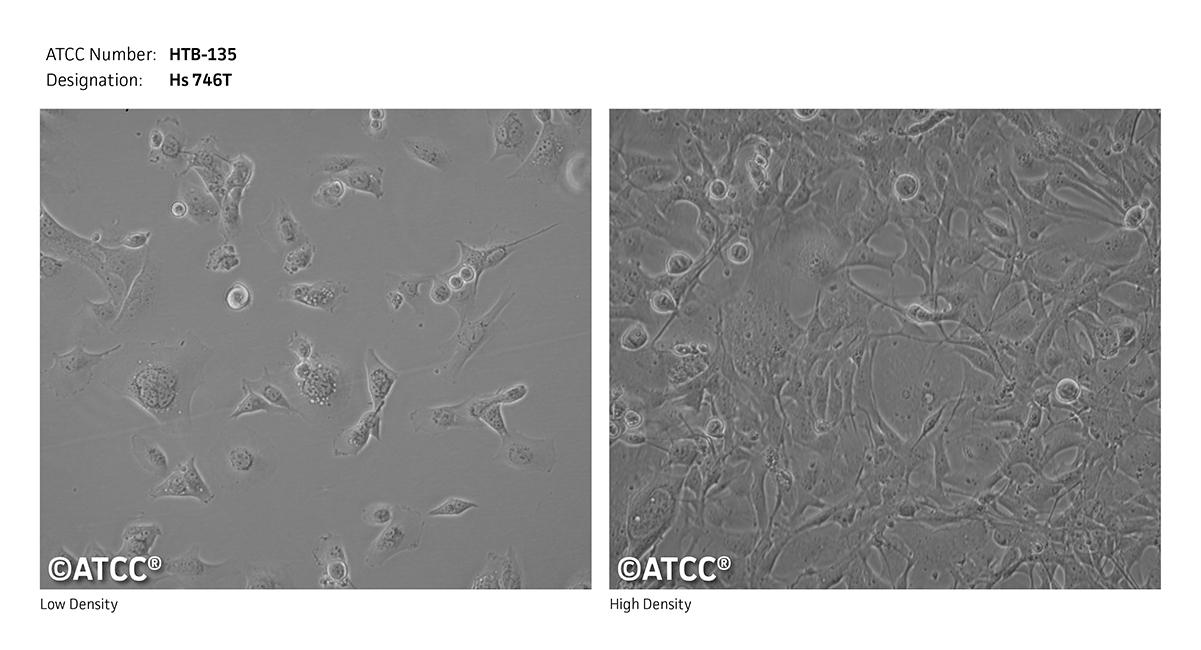 |
Hs 746.T细胞ATCC HTB-135 人胃癌细胞接受后处理
Hs 746.T细胞ATCC HTB-135 人胃癌细胞培养操作
3. 按 6-8ml/瓶补加培养基,轻轻打匀后吸出,在 1000RPM 条件下离心 4 分 钟,弃去上清 液,补加 1-2mL 培养液后吹匀。








