万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 库存:
咨询
- 英文名:
L4440
- CAS号:
咨询
- 保质期:
咨询
- 供应商:
武汉华尔纳生物科技有限公司
- 保存条件:
咨询

- 保存条件:负20度
- 保质期:2年
- 供应商:华纳生物
- 规格:2ug
基本信息
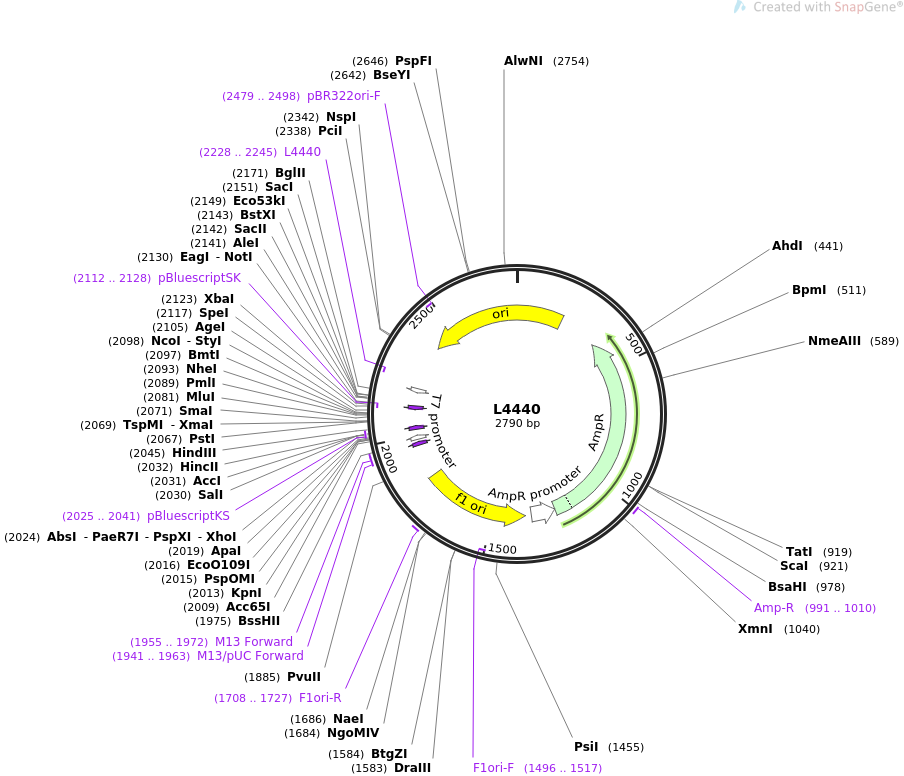
| 启动子: | T7 |
|---|---|
| 复制子: | pUC ori,F1 ori |
| 质粒分类: | 线虫RNAI干扰载体;Worm Expression RNAi |
| 质粒大小: | 2790bp |
| 质粒标签: | lacZN, OriF1 |
| 原核抗性: | 氨苄青霉素Ampicillin |
| 克隆菌株: | DH5α |
| 培养条件: | 37℃,有氧,LB |
| 表达宿主: | 线虫,worm |
| 诱导方式: | IPTG |
| 5'测序引物: | 根据全序列设计 |
质粒简介
Reverse Genetics and RNAi:
RNAi is a natural cellular process in many eukaryotes which scientists have taken advantage of in the lab as a valuable reverse genetics mechanism for regulating gene expression. RNAi involves double-stranded RNA (dsRNA) interfering with the expression of genes with sequences complementary to the dsRNA. By administering a specific dsRNA to a cell culture or multicellular organism, RNAi can be induced to selectively reduce expression of that specific gene in the cells or organism.
RNAi has been particularly well studied in C. elegans, in which the RNAi gene silencing phenotype is heritable and the dsRNA is easily administered. E. coli bacteria carrying RNAi plasmids that produce the desired dsRNA can be fed to worms, and the dsRNA is transferred to the worm via the intestinal tract. RNAi plasmids typically consist of DNA coding sequence from the intended target gene cloned between two T7lac promoters. The plasmid also has a selectable marker that confers resistance to an antibiotic, in this case ampicillin. In 7.15, you will use the E. coli strain HT115 carrying various L4440 plasmids, each containing a different cloned gene sequence. HT115 is an RNase III-deficient E. coli strain with IPTG-inducible T7 polymerase activity. To induce dsRNA production from these plasmids, the HT115 bacteria is grown on special RNAi NGM feeding plates that contain IPTG and the ampicillin analog carbenicillin. Carbenicillin is preferred over ampicillin because it tends to be more stable.
Protocol
A. Preparing feeding plates
1) Inocluate 3mL of LB containing 50 µg/mL ampicillin with individual desired bacterial strain. Grow overnight at 37oC.
2) Seed NGM agar feeding plates (containing 25 µg/mL carbenicillin and 1mM IPTG) by pipetting 150 µL of LB bacterial culture into the center of the plate. You should have three RNAi feeding plates:
a. One plate will be seeded with HT115 bacteria carrying the “empty” L4440 vector (no C. elegans gene is cloned in the vector).
b. Another plate will be seeded with HT115 bacteria carrying a portion of the unc-22 gene cloned into the L4440 vector. The phenotype resulting from RNAi of unc-22 is well-established and is reliably reproducible under the experimental conditions you are using today.
c. A third plate will be seeded with HT115 bacteria carrying a portion of your gene of interest cloned into the L4440 vector. We will use dpy10 as our gene of interest.
3) Let the plates dry overnight at room temperature.
B. Transfer N2 L4 worms
1) Transfer two L4 hermaphrodites from the N2 stock plate to each of the RNAi feeding plates, minimizing the amount of OP50 bacteria transferred. Try to avoid bringing any younger contaminating worms along with the L4 worms you are transferring.
2) Incubate the plates until the progeny reach the desired age (Note: it takes ~4 days for a C. elegans egg to grow into a gravid adult when grown at 16oC.
C. Scoring RNAi phenotypes
1) Observe RNAi controls. Start by looking at the L4440 RNAi plates - what phenotype(s) would you expect to see on these plates? Next, look at the unc-22 RNAi plates – what phenotype(s) would you expect to see on these plates? Record all your observations in your notebook. If you do not see the expected phenotypes on your control plates, this may indicate your RNAi experiment was set-up incorrectly.
2) Observe the RNAi phenotypes for the experimental RNAi construct and record all your observations in your notebook. Examine how the observed phenotypes compare with the corresponding null mutant phenotypes in the same gene (the WormBase database has information on previously characterized RNAi and null mutant phenotypes).
质粒图谱
质粒序列
LOCUS Exported 2790 bp ds-DNA circular SYN 04-SEP-2016
DEFINITION synthetic circular DNA
ACCESSION .
VERSION .
KEYWORDS L4440
SOURCE synthetic DNA construct
ORGANISM synthetic DNA construct
REFERENCE 1 (bases 1 to 2790)
AUTHORS .
TITLE Direct Submission
JOURNAL Exported Sunday, September 4, 2016 from SnapGene Viewer 3.1.4
FEATURES Location/Qualifiers
source 1..2790
/organism="synthetic DNA construct"
/mol_type="other DNA"
promoter 19..37
/note="T7 promoter"
/note="promoter for bacteriophage T7 RNA polymerase"
primer_bind 95..111
/note="SK primer"
/note="common sequencing primer, one of multiple similar
variants"
primer_bind complement(181..197)
/note="KS primer"
/note="common sequencing primer, one of multiple similar
variants"
promoter complement(223..241)
/note="T7 promoter"
/note="promoter for bacteriophage T7 RNA polymerase"
primer_bind complement(251..267)
/note="M13 fwd"
/note="common sequencing primer, one of multiple similar
variants"
rep_origin 409..864
/direction=RIGHT
/note="f1 ori"
/note="f1 bacteriophage origin of replication; arrow
indicates direction of (+) strand synthesis"
promoter 890..994
/gene="bla"
/note="AmpR promoter"
CDS 995..1855
/codon_start=1
/gene="bla"
/product="beta-lactamase"
/note="AmpR"
/note="confers resistance to ampicillin, carbenicillin, and
related antibiotics"
/translation="MSIQHFRVALIPFFAAFCLPVFAHPETLVKVKDAEDQLGARVGYI
ELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRIDAGQEQLGRRIHYSQNDLVEYS
PVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRLDRW
EPELNEAIPNDERDTTMPVAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPLLRSA
LPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIAEIGAS
LIKHW"
rep_origin 2026..2614
/direction=RIGHT
/note="pUC ori"
/note="high-copy-number ColE1/pMB1/pBR322/pUC origin of
replication"
ORIGIN
1 aacctggctt atcgaaatta atacgactca ctatagggag accggcagat ctgatatcat
61 cgatgaattc gagctccacc gcggtggcgg ccgctctaga actagtggat ccaccggttc
121 catggctagc cacgtgacgc gtggatcccc cgggctgcag gaattcgata tcaagcttat
181 cgataccgtc gacctcgagg gggggcccgg tacccaattc gccctatagt gagtcgtatt
241 acgcgcgctc actggccgtc gttttacaac gtcgtgactg ggaaaaccct ggcgttaccc
301 aacttaatcg ccttgcagca catccccctt tcgccagctg gcgtaatagc gaagaggccc
361 gcaccgatcg cccttcccaa cagttgcgca gcctgaatgg cgaatgggac gcgccctgta
421 gcggcgcatt aagcgcggcg ggtgtggtgg ttacgcgcag cgtgaccgct acacttgcca
481 gcgccctagc gcccgctcct ttcgctttct tcccttcctt tctcgccacg ttcgccggct
541 ttccccgtca agctctaaat cgggggctcc ctttagggtt ccgatttagt gctttacggc
601 acctcgaccc caaaaaactt gattagggtg atggttcacg tagtgggcca tcgccctgat
661 agacggtttt tcgccctttg acgttggagt ccacgttctt taatagtgga ctcttgttcc
721 aaactggaac aacactcaac cctatctcgg tctattcttt tgatttataa gggattttgc
781 cgatttcggc ctattggtta aaaaatgagc tgatttaaca aaaatttaac gcgaatttta
841 acaaaatatt aacgcttaca atttaggtgg cacttttcgg ggaaatgtgc gcggaacccc
901 tatttgttta tttttctaaa tacattcaaa tatgtatccg ctcatgagac aataaccctg
961 ataaatgctt caataatatt gaaaaaggaa gagtatgagt attcaacatt tccgtgtcgc
1021 ccttattccc ttttttgcgg cattttgcct tcctgttttt gctcacccag aaacgctggt
1081 gaaagtaaaa gatgctgaag atcagttggg tgcacgagtg ggttacatcg aactggatct
1141 caacagcggt aagatccttg agagttttcg ccccgaagaa cgttttccaa tgatgagcac
1201 ttttaaagtt ctgctatgtg gcgcggtatt atcccgtatt gacgccgggc aagagcaact
1261 cggtcgccgc atacactatt ctcagaatga cttggttgag tactcaccag tcacagaaaa
1321 gcatcttacg gatggcatga cagtaagaga attatgcagt gctgccataa ccatgagtga
1381 taacactgcg gccaacttac ttctgacaac gatcggagga ccgaaggagc taaccgcttt
1441 tttgcacaac atgggggatc atgtaactcg ccttgatcgt tgggaaccgg agctgaatga
1501 agccatacca aacgacgagc gtgacaccac gatgcctgta gcaatggcaa caacgttgcg
1561 caaactatta actggcgaac tacttactct agcttcccgg caacaattaa tagactggat
1621 ggaggcggat aaagttgcag gaccacttct gcgctcggcc cttccggctg gctggtttat
1681 tgctgataaa tctggagccg gtgagcgtgg gtctcgcggt atcattgcag cactggggcc
1741 agatggtaag ccctcccgta tcgtagttat ctacacgacg gggagtcagg caactatgga
1801 tgaacgaaat agacagatcg ctgagatagg tgcctcactg attaagcatt ggtaactgtc
1861 agaccaagtt tactcatata tactttagat tgatttaaaa cttcattttt aatttaaaag
1921 gatctaggtg aagatccttt ttgataatct catgaccaaa atcccttaac gtgagttttc
1981 gttccactga gcgtcagacc ccgtagaaaa gatcaaagga tcttcttgag atcctttttt
2041 tctgcgcgta atctgctgct tgcaaacaaa aaaaccaccg ctaccagcgg tggtttgttt
2101 gccggatcaa gagctaccaa ctctttttcc gaaggtaact ggcttcagca gagcgcagat
2161 accaaatact gtccttctag tgtagccgta gttaggccac cacttcaaga actctgtagc
2221 accgcctaca tacctcgctc tgctaatcct gttaccagtg gctgctgcca gtggcgataa
2281 gtcgtgtctt accgggttgg actcaagacg atagttaccg gataaggcgc agcggtcggg
2341 ctgaacgggg ggttcgtgca cacagcccag cttggagcga acgacctaca ccgaactgag
2401 atacctacag cgtgagctat gagaaagcgc cacgcttccc gaagggagaa aggcggacag
2461 gtatccggta agcggcaggg tcggaacagg agagcgcacg agggagcttc cagggggaaa
2521 cgcctggtat ctttatagtc ctgtcgggtt tcgccacctc tgacttgagc gtcgattttt
2581 gtgatgctcg tcaggggggc ggagcctatg gaaaaacgcc agcaacgcgg cctttttacg
2641 gttcctggcc ttttgctggc cttttgctca catgttcttt cctgcgttat cccctgattc
2701 tgtggataac cgtattaccg cctttgagtg agctgatacc gctcgccgca gccgaacgac
2761 cgagcgcagc gagtcagtga gcgaggaagc
 武汉华尔纳生物科技有限公司,简称华尔纳生物(Warner Bio),坐 落于武汉生物产业基地·光谷生物城,是一家生物医学资源开发,科研生 产,产品销售集一体的科技型企业。
武汉华尔纳生物科技有限公司,简称华尔纳生物(Warner Bio),坐 落于武汉生物产业基地·光谷生物城,是一家生物医学资源开发,科研生 产,产品销售集一体的科技型企业。
华尔纳生物(Warner Bio)始终专注于生命科学领域及细胞实验培养 方向,坚持为客户打造优质高效,安全可靠的生命科学产品,我们始终坚 持于以客户为中心、以品质求生存、以价值求发展、以服务求口碑的企业 理念,从而获得全国广大科研用户的一致认可。
团队以高效的服务品质,专业卓越的技术能力构建了华中地区最大的 生物细胞资源典藏中心,经过多年不懈的努力与发展,现与武汉大学、华 中科技大学、武汉协和医院、武汉同济医院、四川大学华西医院、首都医 科大学、中山大学附属第三医院等全国多所高校、医院、研究所单位达成 长期稳定的合作,同时为科研人员提供免费专业的售前售中售后服务,为 您的科研实验保驾护航。

风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验【共享】RNAi实验的网站 This Caenorhabditis elegans RNAi feeding library
were PCR amplified using Research Genetics GenePairs, cloned into the EcoRV site of vector L4440 from Timmons and Fire (Nature, 395, 854) and transformed into bacterial strain HT115 (Gene, 263, 103-112) as described (Nature 408, 325-330). The whole genome
你还在用小白鼠做研究?Nature Methods 奇葩模式生物大盘点
in Microcebus murinus Reveal Size-Invariant Design Principles in Primate Visual Cortex 为题 [4],刊登在 Current Biology 之上。该研究表明体型与啮齿类动物相似的倭狐猴,更适合被用来探究大脑的奥秘,作为「模式动物」而言大有可为。 线虫 2.0 - 诺奖得主的选择 2002 年,在斯德哥尔摩诺贝尔奖医学奖的颁奖典礼上,新晋诺奖得主 John E. Sulston 讲起了他和线虫的不解情缘。 2002 年研究线虫获得
Clone information and RNAi Feeding Protocol
AHRINGER LAB Clone information and RNAi Feeding Protocol Vector and inserts: Genomic fragments obtained by PCR were cloned into the Timmons and Fire feeding vector (L4440
 技术资料
技术资料暂无技术资料 索取技术资料










