相关产品推荐更多 >
万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 应用范围:
产品信息以Bioss网站为准
- 规格:
50ul/100ul
| 规格: | 50ul | 产品价格: | ¥1400.0 |
|---|---|---|---|
| 规格: | 100ul | 产品价格: | ¥2500.0 |
| 产品编号 | bsm-52110R |
| 英文名称 | Histone H3.3 Recombinant Rabbit mAb |
| 中文名称 | Histone H3.3重组兔单抗 |
| 英文别名 | H3 histone family 3A; H3 histone family 3B; H3 histone, family 3B(H3.3B); H3.3; H3.3A; H3.3B; H33_HUMAN; H3F3; H3F3A; H3f3b; Histone H3.3Q; Histone H3.A; Histone H3.B; MGC87782; MGC87783. |
| 产品应用 | WB=1:200-1000, IHC-P=1:100-400, IHC-F=1:100-400, ICC/IF=1:100-500, IF=1:100-500, ELISA=1:5000-10000 Not yet tested in other applications. |
| 交叉反应 | Human |
| 抗体来源 | Rabbit |
| 免疫原 | Recombinant human Histone H3.3 protein, full length. |
| 亚型 | IgG |
| 性状 | Liquid |
| 纯化方法 | affinity purified by Protein A |
| 克隆类型 | Recombinant |
| 理论分子量 | 15 kDa |
| 浓度 | 1mg/ml |
| 储存液 | 0.01M TBS (pH7.4) with 1% BSA, 0.02% Proclin300 and 50% Glycerol. |
| 研究领域 | Epigenetics and Nuclear Signaling > ChIP assays > ChIP antibodies Epigenetics and Nuclear Signaling > Histones > Variants |
| 亚细胞定位 | Nucleus. Chromosome. |
| 翻译后修饰 | Acetylation is generally linked to gene activation. Acetylation on Lys-10 (H3K9ac) impairs methylation at Arg-9 (H3R8me2s). Acetylation on Lys-19 (H3K18ac) and Lys-24 (H3K24ac) favors methylation at Arg-18 (H3R17me). Acetylation at Lys-123 (H3K122ac) by EP300/p300 plays a central role in chromatin structure: localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability. Citrullination at Arg-9 (H3R8ci) and/or Arg-18 (H3R17ci) by PADI4 impairs methylation and represses transcription. Asymmetric dimethylation at Arg-18 (H3R17me2a) by CARM1 is linked to gene activation. Symmetric dimethylation at Arg-9 (H3R8me2s) by PRMT5 is linked to gene repression. Asymmetric dimethylation at Arg-3 (H3R2me2a) by PRMT6 is linked to gene repression and is mutually exclusive with H3 Lys-5 methylation (H3K4me2 and H3K4me3). H3R2me2a is present at the 3' of genes regardless of their transcription state and is enriched on inactive promoters, while it is absent on active promoters. Specifically enriched in modifications associated with active chromatin such as methylation at Lys-5 (H3K4me), Lys-37 and Lys-80. Methylation at Lys-5 (H3K4me) facilitates subsequent acetylation of H3 and H4. Methylation at Lys-80 (H3K79me) is associated with DNA double-strand break (DSB) responses and is a specific target for TP53BP1. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me), which are linked to gene repression, are underrepresented. Methylation at Lys-10 (H3K9me) is a specific target for HP1 proteins (CBX1, CBX3 and CBX5) and prevents subsequent phosphorylation at Ser-11 (H3S10ph) and acetylation of H3 and H4. Methylation at Lys-5 (H3K4me) and Lys-80 (H3K79me) require preliminary monoubiquitination of H2B at 'Lys-120'. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are enriched in inactive X chromosome chromatin. Monomethylation at Lys-57 (H3K56me1) by EHMT2/G9A in G1 phase promotes interaction with PCNA and is required for DNA replication. Phosphorylated at Thr-4 (H3T3ph) by HASPIN during prophase and dephosphorylated during anaphase. Phosphorylation at Ser-11 (H3S10ph) by AURKB is crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis. In addition phosphorylation at Ser-11 (H3S10ph) by RPS6KA4 and RPS6KA5 is important during interphase because it enables the transcription of genes following external stimulation, like mitogens, stress, growth factors or UV irradiation and result in the activation of genes, such as c-fos and c-jun. Phosphorylation at Ser-11 (H3S10ph), which is linked to gene activation, prevents methylation at Lys-10 (H3K9me) but facilitates acetylation of H3 and H4. Phosphorylation at Ser-11 (H3S10ph) by AURKB mediates the dissociation of HP1 proteins (CBX1, CBX3 and CBX5) from heterochromatin. Phosphorylation at Ser-11 (H3S10ph) is also an essential regulatory mechanism for neoplastic cell transformation. Phosphorylated at Ser-29 (H3S28ph) by MAP3K20 isoform 1, RPS6KA5 or AURKB during mitosis or upon ultraviolet B irradiation. Phosphorylation at Thr-7 (H3T6ph) by PRKCB is a specific tag for epigenetic transcriptional activation that prevents demethylation of Lys-5 (H3K4me) by LSD1/KDM1A. At centromeres, specifically phosphorylated at Thr-12 (H3T11ph) from prophase to early anaphase, by DAPK3 and PKN1. Phosphorylation at Thr-12 (H3T11ph) by PKN1 is a specific tag for epigenetic transcriptional activation that promotes demethylation of Lys-10 (H3K9me) by KDM4C/JMJD2C. Phosphorylation at Tyr-42 (H3Y41ph) by JAK2 promotes exclusion of CBX5 (HP1 alpha) from chromatin. Phosphorylation on Ser-32 (H3S31ph) is specific to regions bordering centromeres in metaphase chromosomes. Ubiquitinated. Monoubiquitinated by RAG1 in lymphoid cells, monoubiquitination is required for V(D)J recombination. Lysine deamination at Lys-5 (H3K4all) to form allysine is mediated by LOXL2. Allysine formation by LOXL2 only takes place on H3K4me3 and results in gene repression. rotonylation (Kcr) is specifically present in male germ cells and marks testis-specific genes in post-meiotic cells, including X-linked genes that escape sex chromosome inactivation in haploid cells. Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors. It is also associated with post-meiotically activated genes on autosomes. Butyrylation of histones marks active promoters and competes with histone acetylation. It is present during late spermatogenesis. Succinylation at Lys-80 (H3K79succ) by KAT2A takes place with a maximum frequency around the transcription start sites of genes (PubMed:29211711). It gives a specific tag for epigenetic transcription activation (PubMed:29211711). |
| 相似性 | Belongs to the histone H3 family. |
| 功能 | Variant histone H3 which replaces conventional H3 in a wide range of nucleosomes in active genes. Constitutes the predominant form of histone H3 in non-dividing cells and is incorporated into chromatin independently of DNA synthesis. Deposited at sites of nucleosomal displacement throughout transcribed genes, suggesting that it represents an epigenetic imprint of transcriptionally active chromatin. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. |
| 保存条件 | Shipped at 4℃. Store at -20℃ for one year. Avoid repeated freeze/thaw cycles. |
| 注意事项 | This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |
| 背景资料 | The nucleosome is a histone octamer containing two molecules each of H2A, H2B, H3 and H4 assembled in one H3-H4 heterotetramer and two H2A-H2B heterodimers. The octamer wraps approximately 147 bp of DNA. Interacts with HIRA, a chaperone required for its incorporation into nucleosomes. Interacts with ZMYND11; when trimethylated at 'Lys-36' (H3.3K36me3). |
| 应用 | 推荐稀释比例 |
| {WB} | {1:200-1000} |
| {IHC-P} | {1:100-400} |
| {IHC-F} | {1:100-400} |
| {ICC/IF} | {1:100-500} |
| {IF} | {1:100-500} |
| {ELISA} | {1:5000-10000} |
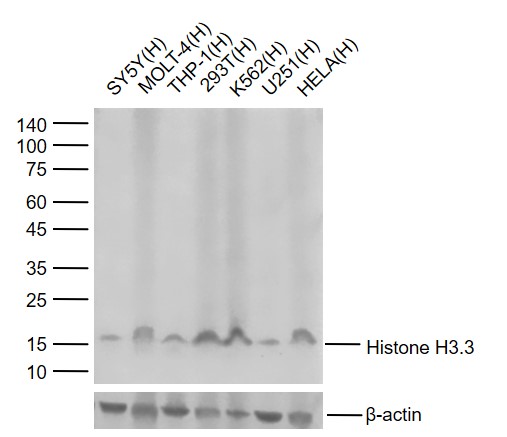
Lane 1: Human SY5Y cell lysates
Lane 2: Human MOLT-4 cell lysates
Lane 3: Human THP-1 cell lysates
Lane 4: Human 293T Cell Lysates
Lane 5: Human K562 Cell Lysates
Lane 6: Human U251 Cell Lysates
Lane 7: Human HELA Cell Lysates
Primary: Anti-Histone H3.3 (15kD) (bsm-52110R) at 1/1000 dilution
Secondary: IRDye800CW Goat Anti-Rabbit IgG at 1/20000 dilution
Predicted band size: 15 kDa
Observed band size: 15 kDa
风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验[IF={{ 2.8 }}] {Zeng Yan. et al. Carvedilol induces pyroptosis through NLRP3-caspase1-ASC inflammasome by nuclear migration of NF-κB in prostate cancer models. MOL BIOL REP. 2024 Dec;51(1):1-12} {WB} {Human}
of both recombinant and native chromatin complexes consisting either of histone subunits alone or in association with accessory proteins, in this case histone chaperones. The approaches described may be generally applicable for monitoring the interactions of a diverse
Competing Chromosomal Proteins from Drosophila Polytene Chromosomes Using Modified Histone Peptides
to chromatin, by its histone tail interaction or by other interactions, then you would not expect to see loss of binding upon peptide competition. A good positive control for the assay is to use H3K9 dimethylated peptide to compete HP1 from the pericentric
INTRODUCTION In cells and tissues, the histone proteins that constitute the nucleosomes can present multiple post-translational modifications, such as lysine acetylation, lysine and arginine methylation, serine phosphorylation
 技术资料
技术资料暂无技术资料 索取技术资料








