相关产品推荐更多 >
万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 询价记录
- 文献和实验
- 技术资料
- 库存:
99
- 供应商:
北京义翘神州科技股份有限公司
- 英文名:
SMM 293-TII Expression Medium (Serum free, complete medium)
- 规格:
1L
SMM 293-TII无血清培养基
产品描述:
| SMM 293-TII培养基是专门用于HEK293细胞悬浮培养和瞬时转染的新一代无血清培养基,细胞培养性能和瞬时转染后的重组蛋白产量均明显优于SMM 293-TI及其他同类产品。产品无血清,无抗生素,无动物源性组分,有利于建立稳定的HEK293细胞表达系统。在转染后的表达阶段同时配套使用加料液SMS 293-SUPI能够实现细胞的长时间培养,获得更高的蛋白产量。 |
产品特点:
| • HEK293细胞瞬时转染专用培养基 | • 即用型完全培养基,不用添加其他组分 |
| • 转染前后无需更换培养基 | • 无动物源组分、无血清、无抗生素 |
| • 适用于瞬时转染和稳定株转染以及培养 | • 适用于细胞的悬浮培养和大规模的反应器培养 |
| • 细胞生长密度及重组蛋白产量明显高于同类产品 |
产品参数:
| 外观 | 澄清红色溶液 | 适用细胞株 | HEK293EBNA及相关的细胞(如HEK293F、HEK293E、 HEK293H 、HEK293S、HEK293FT) |
| 渗透压 | 275±10 mOsm/kg | 细胞接种密度 | 0.3-0.4×106 cells/mL |
| pH值 | 6.9–7.4 | 倍增时间 | < 26 hrs(因细胞类型、状态影响而存在数值变化) |
| 无菌检测 | 无菌 | 细胞培养评价 | 优良 |
| 内毒素 | < 10 EU/mL | 产品用途 | 仅用于科学研究,不推荐用于人类或动物的诊断和治疗 |
| 有效期 | 6个月 | 储存条件 | 2-8 ℃,避光 |
SMM 293-TII培养基为澄清红色透明液体,2~8℃避光保存,使用过程中请勿反复预热。
产品使用效果: 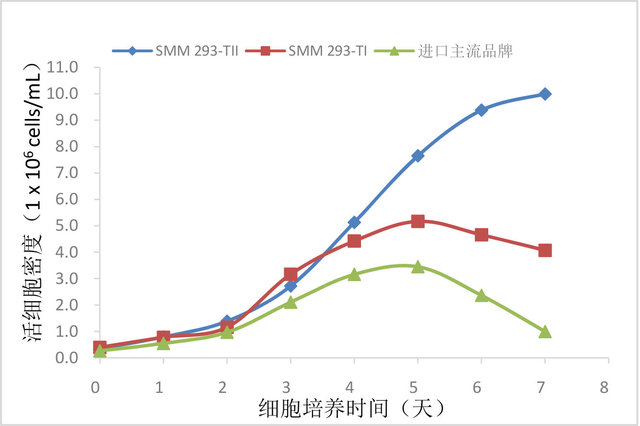
图1. SMM 293-TII生长性能检测。相同培养条件下,检测SMM 293-TII与SMM 293-TI及知名进口品牌HEK293
细胞无血清培养基中生长的HEK293F细胞密度随培养时间的变化。 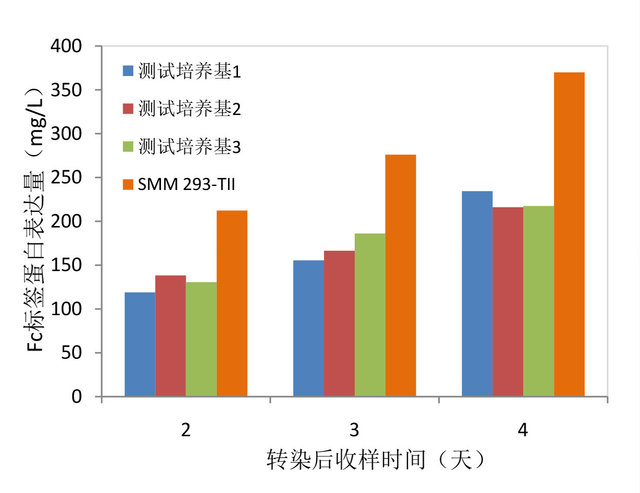
图2. SMM 293-TII蛋白表达量检测。使用相同的Fc标签重组表达质粒转染培养在SMM 293-TII和测其它品牌同
类无血清培养基中的HEK293F,在转染后不同时间收样检测细胞的蛋白产量。
风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
- 作者
- 内容
- 询问日期
 文献和实验
文献和实验1, Guo S, et al. Dosing interval regimen shapes potency and breadth of antibody repertoire after vaccination of SARS-CoV-2 RBD protein subunit vaccine.Cell discovery, PubMed ID: 37507370
2, Zhang Y, et al. Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.Cell research, PubMed ID: 37491604
3, Zhao Z, et al. Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.Nature communications, PubMed ID: 37479708
4, Yao W, et al. Evolution of SARS-CoV-2 Spikes shapes their binding affinities to animal ACE2 orthologs.Microbiology spectrum, PubMed ID: 37943512
5, Liu Y, et al. CTHRC1 promotes colorectal cancer progression by recruiting tumor-associated macrophages via up-regulation of CCL15.Journal of molecular medicine (Berlin, Germany), PubMed ID: 37987774
6, Shi K, et al. Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.mBio, PubMed ID: 38112468
7, Mao YX, et al. Transport mechanism of human bilirubin transporter ABCC2 tuned by the inter-module regulatory domain.Nature communications, PubMed ID: 38316776
8, Li R, et al. Double-layered N-S1 protein nanoparticle immunization elicits robust cellular immune and broad antibody responses against SARS-CoV-2.Journal of nanobiotechnology, PubMed ID: 38291444
9, Ying W, et al. Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.Science (New York, N.Y.), PubMed ID: 38513023
10, Li W, et al. Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.The EMBO journal, PubMed ID: 38467833
11, Cong J, et al. Bile acids modified by the intestinal microbiota promote colorectal cancer growth by suppressing CD8+ T cell effector functions.Immunity, PubMed ID: 38479384
12, Hu S, et al. Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.Structure (London, England : 1993), PubMed ID: 38626767
13, Luo J, et al. Capturing acyl-enzyme intermediates with genetically encoded 2,3-diaminopropionic acid for hydrolase substrate identification.Nature protocols, PubMed ID: 38867073
14, Chen J, et al. The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.Virologica Sinica, PubMed ID: 38866203
15, Yang Q, et al. Farnesyltransferase inhibitor lonafarnib suppresses respiratory syncytial virus infection by blocking conformational change of fusion glycoprotein.Signal transduction and targeted therapy, PubMed ID: 38853183
16, Tong Z, et al. Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.Cell reports, PubMed ID: 38850530
17, Wang H, et al. TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.Cell, PubMed ID: 38964329
18, He J, et al. Structural basis for the transport and substrate selection of human urate transporter 1.Cell reports, PubMed ID: 39146184
19, Li L, et al. Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.Structure (London, England : 1993), PubMed ID: 39013463
20, Chengqi Yi, et al. Protocol for profiling RNA m5C methylation at base resolution using m5C-TAC-seq. STAR protocols, PubMed ID: 39893640
21, Yixiao Zhang, et al. Molecular basis for the stepwise and faithful maturation of the 20S proteasome. Science advances, PubMed ID: 39792683
22, Huiqin Liu, et al. Modulating the complement system through epitope-specific inhibition by complement C3 inhibitors. The Journal of biological chemistry, PubMed ID: 39894217
23, Zexian Zeng, et al. Potentiating anti-tumor immunity by re-engaging immune synapse molecules. Cell reports. Medicine, PubMed ID: 39999838
24, Huan Yan, et al. ACE2 utilization of HKU25 clade MERS-related coronaviruses with broad geographic distribution. bioRxiv : the preprint server for biology, PubMed ID: 40027745
25, Zengqin Deng, et al. Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses. Cell, PubMed ID: 39922191
26, Huan Yan, et al. Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses. Cell, PubMed ID: 39922192
27, Xiaoqian Hu, et al. The HAVCR1-centric host factor network drives Zika virus vertical transmission. Cell reports, PubMed ID: 40156834
28, Rebecca Page, et al. Cryo-EM structures of PP2A:B55 with p107 and Eya3 define substrate recruitment. Nature structural & molecular biology, PubMed ID: 40247147
29, Lizhe Zhu, et al. Structural basis of cytokinin transport by the Arabidopsis AZG2. Nature communications, PubMed ID: 40216803
30, Yunbin Ye, et al. Neoepitope BTLAP267L-specific TCR-T cell immunotherapy unlocks precision treatment for hepatocellular carcinoma. Cancer biology & medicine, PubMed ID: 40205806
31, Xin Gong, et al. Mechanisms of aureobasidin A inhibition and drug resistance in a fungal IPC synthase complex. Nature communications, PubMed ID: 40442105
32, Xinzheng Zhang, et al. Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus. Cell reports, PubMed ID: 40402741
33, Shutang Tan, et al. Structural insights into auxin influx mediated by the Arabidopsis AUX1. Cell, PubMed ID: 40378849
34, Feng Han, et al. Proton perception and activation of a proton-sensing GPCR. Molecular cell, PubMed ID: 40215960
35, Xiaohong Qin, et al. Unprocessed BMP9 precursor is an intrinsic antagonist for its active growth factor. Structure (London, England : 1993), PubMed ID: 40412377
36, Yang-Xin Fu, et al. Sequential intranasal booster triggers class switching from intramuscularly primed IgG to mucosal IgA against SARS-CoV-2. The Journal of clinical investigation, PubMed ID: 39808503
Generating stable cell lines in HEK293
Generating stable cell lines in HEK293 Prior to transfection, it is recommended that you linearize your pcDNA gene construct. Linearizing will decrease the likelihood of the vector integrating into the genome in a way
Construction and Characterization of Adenovirus Vectors
stock solution with H2 O and sterilize by autoclaving for 45 min at 121°C. Complete medium Dialysis buffer (10 mM Tris-HCl, pH 8.0) Sterilize by filtration through a 0.2-µm filter. DNase I (Ad) Ethidium bromide
Lonza 12-765Q Pro293S-CDM, for suspension cells, 1L Lonza 12-764Q Pro293A-CDM, for adherent cells, 1L Lonza BE02-030Q ProVero-1 NAO Medium for Vero Cells, 1L Lonza 12-749Q UltraMDCK Serum-free Medium, 1L 杂交瘤细胞培养基 Lonza BE02










