相关产品推荐更多 >
万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 保存条件:
-20℃
- 保质期:
6个月
- 英文名:
pDsRed-Monomer-N In-Fusion Ready
- 库存:
大量
- 供应商:
钰博生物
- 规格:
1ug/5ug
载体基本信息
| 出品公司: | Ybscience |
|---|---|
| 载体名称: | pDsRed-Monomer-N In-Fusion Ready |
| 质粒类型: | 哺乳动物细胞表达载体;荧光报告载体 |
| 高拷贝/低拷贝: | 高拷贝 |
| 克隆方法: | 限制性内切酶,多克隆位点 |
| 启动子: | CMV |
| 载体大小: | 4741 bp |
| 5' 测序引物及序列: | 5’-GTACTGGAACTGGGGGGACAG-3’ |
| 3' 测序引物及序列: | -- |
| 载体标签: | DsRed-Monomer(C-端) |
| 载体抗性: | 卡那霉素 |
| 筛选标记: | 新霉素(Neomycin) |
| 克隆菌株: | DH5α, HB101 |
| 宿主细胞(系): | 常规细胞系,293、CV-1、CHO等 |
| 备注: | pDsRed-Monomer-N In-Fusion Ready载体是荧光报告载体,表达C端DsRed-Monomer融合蛋白; DsRed-Monomer是DsRed的单体突变体,与DsRed相比,编码序列经过了优化,适用于在哺乳动物细胞中的高水平表达。 |
| 稳定性: | 瞬表达 或 稳表达 |
| 组成型/诱导型: | 组成型 |
| 病毒/非病毒: | 非病毒 |
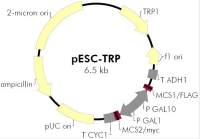
载体质粒图谱和多克隆位点信息



载体简介
pDsRed-Monomer-N In-Fusion Ready载体描述 pDsRed-Monomer-N In-Fusion Ready vector is a linearized mammalian expression vector that encodes DsRed-Monomer (DsRed. M1), a monomeric mutant derived from the tetrameric Discosoma sp. red fluorescent protein, DsRed (1). DsRed-Monomer contains a total of forty-five amino acid substitutions. When DsRed-Monomer is expressed in mammalian cell cultures, red fluorescent cells can be detected by either fluorescence microscopy or flow cytometry 12–16 hr after transfection. DsRed-Monomer has an excitation maximum at 557 nm and an emission maximum at 592 nm. The DsRed-Monomer coding sequence is human codon-optimized for high expression levels in mammalian cells (2).The linearized vector allows direct cloning of PCR products without any need for restriction digestion when using the In-Fusion HD Cloning Plus (638909). This is accomplished by the use of a specific 15 nucleotide long sequence within the 5’ ends of the sense and antisense primers that overlap with the cut ends of the linearized vector. The primers that will be used to amplify In-Fusion Ready PCR products require the following 15 nucleotides on their 5’ends: Sense primer: 5’ AAGGCCTCTGTCGAC followed by sequence of amplification target 3’ Antisense primer: 5’ AGAATTCGCAAGCTT followed by sequence of amplification target 3’ If the sequence of the gene of interest is added in-frame immediately after the 15 nucleotides mentioned above, this sequence will automatically be in-frame with the DsRed-Monomer sequence downstream and therefore be expressed as a fusion protein to the N-terminus of DsRed-Monomer. A Kozak consensus translation initiation site upstream of DsRed-Monomer increases the translation efficiency in eukaryotic cells (3). SV40 polyadenylation signals downstream of the DsRed-Monomer gene direct proper processing of the 3' end of the mRNA. transcript. The vector backbone contains an SV40 origin of replication in mammalian cells expressing the SV40 T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin-resistance cassette (Neor) allows selection of stably transfected eukaryotic cells using G418. This cassette contains the SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene. A bacterial promoter upstream of the cassette allows the plasmid to confer kanamycin resistance in E. coli. Fusions to the N-terminus of DsRed-Monomer retain the fluorescent properties of the native protein and allow monitoring of the fusion protein localization in vivo. The PCR-amplified gene of interest is directly cloned into the pDsRed-Monomer-N In-Fusion Ready Vector so that it is in-frame with the DsRed-Monomer coding sequences with no intervening in-frame stop codons. This can be accomplished by using the suggested primers. The inserted gene must include the initiating ATG codon. This recombinant vector can be transfected into mammalian cells using any standard transfection method. Stable transformants can be selected using G418 (2). The DsRed1-N Sequencing Primer (Cat. No. 632387) can be used to sequence genes cloned adjacent to the 5’ end of the DsRed-Monomer coding region. For Western blotting, the Living Colors DsRed Polyclonal Antibody (Cat. No. 632496) can be used to recognize the DsRed-Monomer protein. For optimal results for immunoprecipitation, we suggest you titrate the amount of antibody needed for quantitative recovery of the antigen. As a starting point, however, we recommend using the antibody at a 1:1,000 dilution. Propagation in E. coli Suitable host strains: Stellar Competent Cells. Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts. E. coli replication origin: pUC Copy number: ~ 500 Plasmid incompatibility group: pMB1/ColE1 Excitation and emission maxima of DsRed-Monomer Excitation maximum = 557 nm Emission maximum = 592 nm
载体序列
LOCUS pDsRed-Monomer-N In-Fusion Ready 4655 bp DNA SYN
DEFINITION pDsRed-Monomer-N In-Fusion Ready
ACCESSION
KEYWORDS
SOURCE
ORGANISM other sequences; artificial sequences; vectors.
FEATURES Location/Qualifiers
source 1..4655
/organism="pDsRed-Monomer-N In-Fusion Ready"
/mol_type="other DNA"
gene 30..701
/label="DsRed_Monomer"
/gene="DsRed_Monomer"
CDS 60..704
/label="ORF frame 3"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
misc_feature complement(207..227)
/label="dsRed1_N_primer"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
misc_feature 923..942
/label="EBV_rev_primer"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
rep_origin complement(1092..1398)
/label="f1_origin"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
promoter 1477..1505
/label="AmpR_promoter"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
misc_feature complement(1571..1591)
/label="pBABE_3_primer"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
misc_feature complement(1577..1792)
/label="SV40_enhancer"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
promoter 1589..1857
/label="SV40_promoter"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
rep_origin 1756..1833
/label="SV40_origin"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
misc_feature 1818..1837
/label="SV40pro_F_primer"
/translation="MQFKVRMEGSVNGHYFEIEGEGEGKPYEGTQTAKLQVTKGGPLP
FAWDILSPQFQYGSKAYVKHPADIPDYMKLSFPEGFTWERSMNFEDGGVVEVQQDSSL
QDGTFIYKVKFKGVNFPADGPVMQKKTAGWEPSTEKLYPQDGVLKGEISHALKLKDGG
HYTCDFKTVYKAKKPVQLPGNHYVDSKLDITNHNEDYTVVEQYEHAEARHSGSQ*"
CDS 1940..2734
/label="ORF frame 2"
/translation="MIEQDGLHAGSPAAWVERLFGYDWAQQTIGCSDAAVFRLSAQGR
PVLFVKTDLSGALNELQDEAARLSWLATTGVPCAAVLDVVTEAGRDWLLLGEVPGQDL
LSSHLAPAEKVSIMADAMRRLHTLDPATCPFDHQAKHRIERARTRMEAGLVDQDDLDE
EHQGLAPAELFARLKASMPDGEDLVVTHGDACLPNIMVENGRFSGFIDCGRLGVADRY
QDIALATRDIAEELGGEWADRFLVLYGIAAPDSQRIAFYRLLDEFF*"
gene 1943..2731
/label="NeoR/KanR"
/gene="NeoR/KanR"
/translation="MIEQDGLHAGSPAAWVERLFGYDWAQQTIGCSDAAVFRLSAQGR
PVLFVKTDLSGALNELQDEAARLSWLATTGVPCAAVLDVVTEAGRDWLLLGEVPGQDL
LSSHLAPAEKVSIMADAMRRLHTLDPATCPFDHQAKHRIERARTRMEAGLVDQDDLDE
EHQGLAPAELFARLKASMPDGEDLVVTHGDACLPNIMVENGRFSGFIDCGRLGVADRY
QDIALATRDIAEELGGEWADRFLVLYGIAAPDSQRIAFYRLLDEFF*"
CDS complement(2249..2785)
/label="ORF frame 2"
/translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA
VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA
VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ
PPHCISHDGYFLGRSKVR*"
terminator 2909..3178
/label="TK_PA_terminator"
/translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA
VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA
VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ
PPHCISHDGYFLGRSKVR*"
rep_origin 3326..3945
/label="pBR322_origin"
/translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA
VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA
VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ
PPHCISHDGYFLGRSKVR*"
promoter 4054..4606
/label="CMV_immearly_promoter"
/translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA
VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA
VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ
PPHCISHDGYFLGRSKVR*"
misc_feature 4109..4396
/label="CAG_enhancer"
/translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA
VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA
VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ
PPHCISHDGYFLGRSKVR*"
misc_feature 4563..4583
/label="CMV_fwd_primer"
/translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA
VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA
VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ
PPHCISHDGYFLGRSKVR*"
promoter 4564..4633
/label="CMV_promoter"
/translation="MAGWASLGRSFRTPESRSEELVKKAIEGDALRIGSGDTVKHEEA
VSPFAAKLFSNITGSQRYVLIAVRHTQPATVDESRKAAIFHHDIRQAGIAMGHDEILA
VGHARLEPGEQFGWREPLMLFVQIILIDKTGFHPSTCSLDAMFRLVVEWAGSRIKRMQ
PPHCISHDGYFLGRSKVR*"
ORIGIN
1 AGCTTGCGAA TTCTGGCCTA CCGGTCGCGG ACAACACCGA GGACGTCATC AAGGAGTTCA
61 TGCAGTTCAA GGTGCGCATG GAGGGCTCCG TGAACGGCCA CTACTTCGAG ATCGAGGGCG
121 AGGGCGAGGG CAAGCCCTAC GAGGGCACCC AGACCGCCAA GCTGCAGGTG ACCAAGGGCG
181 GCCCCCTGCC CTTCGCCTGG GACATCCTGT CCCCCCAGTT CCAGTACGGC TCCAAGGCCT
241 ACGTGAAGCA CCCCGCCGAC ATCCCCGACT ACATGAAGCT GTCCTTCCCC GAGGGCTTCA
301 CCTGGGAGCG CTCCATGAAC TTCGAGGACG GCGGCGTGGT GGAGGTGCAG CAGGACTCCT
361 CCCTGCAGGA CGGCACCTTC ATCTACAAGG TGAAGTTCAA GGGCGTGAAC TTCCCCGCCG
421 ACGGCCCCGT AATGCAGAAG AAGACTGCCG GCTGGGAGCC CTCCACCGAG AAGCTGTACC
481 CCCAGGACGG CGTGCTGAAG GGCGAGATCT CCCACGCCCT GAAGCTGAAG GACGGCGGCC
541 ACTACACCTG CGACTTCAAG ACCGTGTACA AGGCCAAGAA GCCCGTGCAG CTGCCCGGCA
601 ACCACTACGT GGACTCCAAG CTGGACATCA CCAACCACAA CGAGGACTAC ACCGTGGTGG
661 AGCAGTACGA GCACGCCGAG GCCCGCCACT CCGGCTCCCA GTAGGCGGCC GCGACTGCGA
721 CTCTAGATCA TAATCAGCCA TACCACATTT GTAGAGGTTT TACTTGCTTT AAAAAACCTC
781 CCACACCTCC CCCTGAACCT GAAACATAAA ATGAATGCAA TTGTTGTTGT TAACTTGTTT
841 ATTGCAGCTT ATAATGGTTA CAAATAAAGC AATAGCATCA CAAATTTCAC AAATAAAGCA
901 TTTTTTTCAC TGCATTCTAG TTGTGGTTTG TCCAAACTCA TCAATGTATC TTAAGGCGTA
961 AATTGTAAGC GTTAATATTT TGTTAAAATT CGCGTTAAAT TTTTGTTAAA TCAGCTCATT
1021 TTTTAACCAA TAGGCCGAAA TCGGCAAAAT CCCTTATAAA TCAAAAGAAT AGACCGAGAT
1081 AGGGTTGAGT GTTGTTCCAG TTTGGAACAA GAGTCCACTA TTAAAGAACG TGGACTCCAA
1141 CGTCAAAGGG CGAAAAACCG TCTATCAGGG CGATGGCCCA CTACGTGAAC CATCACCCTA
1201 ATCAAGTTTT TTGGGGTCGA GGTGCCGTAA AGCACTAAAT CGGAACCCTA AAGGGAGCCC
1261 CCGATTTAGA GCTTGACGGG GAAAGCCGGC GAACGTGGCG AGAAAGGAAG GGAAGAAAGC
1321 GAAAGGAGCG GGCGCTAGGG CGCTGGCAAG TGTAGCGGTC ACGCTGCGCG TAACCACCAC
1381 ACCCGCCGCG CTTAATGCGC CGCTACAGGG CGCGTCAGGT GGCACTTTTC GGGGAAATGT
1441 GCGCGGAACC CCTATTTGTT TATTTTTCTA AATACATTCA AATATGTATC CGCTCATGAG
1501 ACAATAACCC TGATAAATGC TTCAATAATA TTGAAAAAGG AAGAGTCCTG AGGCGGAAAG
1561 AACCAGCTGT GGAATGTGTG TCAGTTAGGG TGTGGAAAGT CCCCAGGCTC CCCAGCAGGC
1621 AGAAGTATGC AAAGCATGCA TCTCAATTAG TCAGCAACCA GGTGTGGAAA GTCCCCAGGC
1681 TCCCCAGCAG GCAGAAGTAT GCAAAGCATG CATCTCAATT AGTCAGCAAC CATAGTCCCG
1741 CCCCTAACTC CGCCCATCCC GCCCCTAACT CCGCCCAGTT CCGCCCATTC TCCGCCCCAT
1801 GGCTGACTAA TTTTTTTTAT TTATGCAGAG GCCGAGGCCG CCTCGGCCTC TGAGCTATTC
1861 CAGAAGTAGT GAGGAGGCTT TTTTGGAGGC CTAGGCTTTT GCAAAGATCG ATCAAGAGAC
1921 AGGATGAGGA TCGTTTCGCA TGATTGAACA AGATGGATTG CACGCAGGTT CTCCGGCCGC
1981 TTGGGTGGAG AGGCTATTCG GCTATGACTG GGCACAACAG ACAATCGGCT GCTCTGATGC
2041 CGCCGTGTTC CGGCTGTCAG CGCAGGGGCG CCCGGTTCTT TTTGTCAAGA CCGACCTGTC
2101 CGGTGCCCTG AATGAACTGC AAGACGAGGC AGCGCGGCTA TCGTGGCTGG CCACGACGGG
2161 CGTTCCTTGC GCAGCTGTGC TCGACGTTGT CACTGAAGCG GGAAGGGACT GGCTGCTATT
2221 GGGCGAAGTG CCGGGGCAGG ATCTCCTGTC ATCTCACCTT GCTCCTGCCG AGAAAGTATC
2281 CATCATGGCT GATGCAATGC GGCGGCTGCA TACGCTTGAT CCGGCTACCT GCCCATTCGA
2341 CCACCAAGCG AAACATCGCA TCGAGCGAGC ACGTACTCGG ATGGAAGCCG GTCTTGTCGA
2401 TCAGGATGAT CTGGACGAAG AGCATCAGGG GCTCGCGCCA GCCGAACTGT TCGCCAGGCT
2461 CAAGGCGAGC ATGCCCGACG GCGAGGATCT CGTCGTGACC CATGGCGATG CCTGCTTGCC
2521 GAATATCATG GTGGAAAATG GCCGCTTTTC TGGATTCATC GACTGTGGCC GGCTGGGTGT
2581 GGCGGACCGC TATCAGGACA TAGCGTTGGC TACCCGTGAT ATTGCTGAAG AGCTTGGCGG
2641 CGAATGGGCT GACCGCTTCC TCGTGCTTTA CGGTATCGCC GCTCCCGATT CGCAGCGCAT
2701 CGCCTTCTAT CGCCTTCTTG ACGAGTTCTT CTGAGCGGGA CTCTGGGGTT CGAAATGACC
2761 GACCAAGCGA CGCCCAACCT GCCATCACGA GATTTCGATT CCACCGCCGC CTTCTATGAA
2821 AGGTTGGGCT TCGGAATCGT TTTCCGGGAC GCCGGCTGGA TGATCCTCCA GCGCGGGGAT
2881 CTCATGCTGG AGTTCTTCGC CCACCCTAGG GGGAGGCTAA CTGAAACACG GAAGGAGACA
2941 ATACCGGAAG GAACCCGCGC TATGACGGCA ATAAAAAGAC AGAATAAAAC GCACGGTGTT
3001 GGGTCGTTTG TTCATAAACG CGGGGTTCGG TCCCAGGGCT GGCACTCTGT CGATACCCCA
3061 CCGAGACCCC ATTGGGGCCA ATACGCCCGC GTTTCTTCCT TTTCCCCACC CCACCCCCCA
3121 AGTTCGGGTG AAGGCCCAGG GCTCGCAGCC AACGTCGGGG CGGCAGGCCC TGCCATAGCC
3181 TCAGGTTACT CATATATACT TTAGATTGAT TTAAAACTTC ATTTTTAATT TAAAAGGATC
3241 TAGGTGAAGA TCCTTTTTGA TAATCTCATG ACCAAAATCC CTTAACGTGA GTTTTCGTTC
3301 CACTGAGCGT CAGACCCCGT AGAAAAGATC AAAGGATCTT CTTGAGATCC TTTTTTTCTG
3361 CGCGTAATCT GCTGCTTGCA AACAAAAAAA CCACCGCTAC CAGCGGTGGT TTGTTTGCCG
3421 GATCAAGAGC TACCAACTCT TTTTCCGAAG GTAACTGGCT TCAGCAGAGC GCAGATACCA
3481 AATACTGTTC TTCTAGTGTA GCCGTAGTTA GGCCACCACT TCAAGAACTC TGTAGCACCG
3541 CCTACATACC TCGCTCTGCT AATCCTGTTA CCAGTGGCTG CTGCCAGTGG CGATAAGTCG
3601 TGTCTTACCG GGTTGGACTC AAGACGATAG TTACCGGATA AGGCGCAGCG GTCGGGCTGA
3661 ACGGGGGGTT CGTGCACACA GCCCAGCTTG GAGCGAACGA CCTACACCGA ACTGAGATAC
3721 CTACAGCGTG AGCTATGAGA AAGCGCCACG CTTCCCGAAG GGAGAAAGGC GGACAGGTAT
3781 CCGGTAAGCG GCAGGGTCGG AACAGGAGAG CGCACGAGGG AGCTTCCAGG GGGAAACGCC
3841 TGGTATCTTT ATAGTCCTGT CGGGTTTCGC CACCTCTGAC TTGAGCGTCG ATTTTTGTGA
3901 TGCTCGTCAG GGGGGCGGAG CCTATGGAAA AACGCCAGCA ACGCGGCCTT TTTACGGTTC
3961 CTGGCCTTTT GCTGGCCTTT TGCTCACATG TTCTTTCCTG CGTTATCCCC TGATTCTGTG
4021 GATAACCGTA TTACCGCCAT GCATTAGTTA TTAATAGTAA TCAATTACGG GGTCATTAGT
4081 TCATAGCCCA TATATGGAGT TCCGCGTTAC ATAACTTACG GTAAATGGCC CGCCTGGCTG
4141 ACCGCCCAAC GACCCCCGCC CATTGACGTC AATAATGACG TATGTTCCCA TAGTAACGCC
4201 AATAGGGACT TTCCATTGAC GTCAATGGGT GGAGTATTTA CGGTAAACTG CCCACTTGGC
4261 AGTACATCAA GTGTATCATA TGCCAAGTAC GCCCCCTATT GACGTCAATG ACGGTAAATG
4321 GCCCGCCTGG CATTATGCCC AGTACATGAC CTTATGGGAC TTTCCTACTT GGCAGTACAT
4381 CTACGTATTA GTCATCGCTA TTACCATGGT GATGCGGTTT TGGCAGTACA TCAATGGGCG
4441 TGGATAGCGG TTTGACTCAC GGGGATTTCC AAGTCTCCAC CCCATTGACG TCAATGGGAG
4501 TTTGTTTTGG CACCAAAATC AACGGGACTT TCCAAAATGT CGTAACAACT CCGCCCCATT
4561 GACGCAAATG GGCGGTAGGC GTGTACGGTG GGAGGTCTAT ATAAGCAGAG CTGGTTTAGT
4621 GAACCGTCAG ATCCGCTAGC TAAGGCCTCT GTCGA
风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验is reached (1 hour). REAGENTS USED IN GELS Separating gel monomer (49.5% T 6% C) Mix 93.0 g acrylamide and 6.0 g N, N'' -methylene-bisacrylamide in a total volume of 200 ml dH2O. Filter solution through a 0.45 µm filter
Creating Transient Cell Membrane Pores Using a Standard Inkjet Printer
a) Ensure that the ethanol has dried before adding the bioink printing solution. 4. Making Cell Suspension - "Bioink" 1) Culture cells until ready to passage. a) For 3T3 fibroblasts: Seed cells on T-75
.), which does not seem to have been used as a fixative. Carbodiimides are compounds that combine with and cross-link carboxyl groups. They fix proteins by joining together C-termini and/or side chains of glutamic and aspartic acid units. Their general chemical formula is R-N=C=N-R
 技术资料
技术资料暂无技术资料 索取技术资料