相关产品推荐更多 >
万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 品系:
详见细胞说明资料
- 细胞类型:
详见细胞说明资料
- 肿瘤类型:
详见细胞说明资料
- 供应商:
上海冠导生物工程有限公司
- 库存:
≥100瓶
- 生长状态:
详见细胞说明资料
- 年限:
详见细胞说明资料
- 运输方式:
常温运输【复苏细胞】或干冰运输【冻存细胞】
- 器官来源:
详见细胞说明资料
- 是否是肿瘤细胞:
详见细胞说明资料
- 细胞形态:
详见细胞说明资料
- 免疫类型:
详见细胞说明资料
- 物种来源:
详见细胞说明资料
- 相关疾病:
详见细胞说明资料
- 组织来源:
详见细胞说明资料
- 英文名:
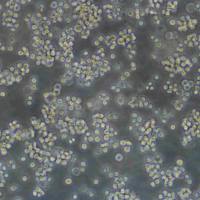
HCT 116人结肠癌传代细胞|送STR图谱
- 规格:
1*10(6)Cellls/瓶
"HCT 116人结肠癌传代细胞|送STR图谱
传代方法:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
换液频率:每周2-3次
背景资料:是一种人结肠癌细胞系,是由M·Brattain等人于1979年从患结肠癌的男性病人中分离的三株恶性细胞中的一株。HCT 116细胞在半固体琼脂糖培养基中形成克隆;HCT 116细胞在无胸腺裸鼠有致瘤性,形成肿瘤结节。
常见细胞贴壁较弱原因:在遇到运输低温及震荡、室温静置太长时间、添加的培养基或其他试剂过冷、密度较高、聚集未吹散、加液吹打到细胞面等情况时会出现明显的成片脱落现象,此时若脱落现象不严重应尽快放回培养基继续培养,若呈大片脱落的情况时需要收集细胞重新消化吹散并接种;建议使用经过包被或者高贴壁培养瓶培养细胞,尽量避免接触低温或密度过高。
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
VP303 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Huh-7 Cells、COLO 205 Cells、MOLP-8 Cells
BT-20 Cells;背景说明:该细胞1958年由E.Y. Lasfargues 和 L. Ozzello 建系,源自一位74岁白人女性的乳腺癌组织。该细胞表达WNT3和WNT78。TNF alpha抑制该细胞生长。该细胞雌激素受体阴性,但表达5'外显子缺失的雌激素mRNA。;传代方法:1:2—1:4传代,2—3天换液一次;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:Ramos (RA 1) Cells、H-1694 Cells、MC-4 Cells
Hx-147 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:PLB 985 Cells、MES-23.5 Cells、PG-4 Cells
133B Cells(拥有STR基因鉴定图谱)
HCT 116人结肠癌传代细胞|送STR图谱
产品包装形式:复苏细胞:T25培养瓶(一瓶)或冻存细胞:1ml冻存管(两支)
来源说明:细胞主要来源ATCC、DSMZ等细胞库
ATCC细胞库(American Type Culture Colection),该中心一直致力于细胞分类、鉴定和保藏工作。ATCC是全球最大的生物资源保藏中心,ATCC通过行业标准产品、服务和创新解决方案支持全球学术、政府、生物技术、制药、食品、农业和工业领域的科学进步。ATCC提供的服务和定制解决方案包括细胞和微生物培养、鉴定、生物衍生物的开发和生产、性能测试和生物资源保藏服务。美国国家标准协会(ANSI)认可了ATCC标准开发组织,并制定了标准协议,以确保生物材料的可靠性和可重复性。ATCC的使命是为了获取、鉴定、保存、开发、标准化和分发生物资源和生物信息,以提高和应用生物科学知识。
物种来源:Human\Mouse\Rat\Others
HCT 116人结肠癌传代细胞|送STR图谱
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
SNUC2B Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:PaTu 8988 S Cells、H-727 Cells、C-28/I2 Cells
SCC-25 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:10传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:ZR75-1 Cells、HUT 28 Cells、RPMI #1846 Cells
D 407 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:HCT 15 Cells、293 HEK Cells、ECC12 Cells
形态特性:上皮细胞样
细胞冻存复苏材料与方法步骤:常用的细胞冷冻贮存器为贮存器,规格有35L和50L两种。使用时要注意以下几点:(1)一般两周需充一次,至少一个月充一次。温度达-196℃,使用时注意勿让溅到皮肤上,以免引起冻伤。(2)容器为双层结构,中间为真空层,瓶口有双层焊接处,应防止焊接部裂开。(3)在装入时,要注意缓慢小心,并用厚纸卷筒或制漏斗作引导,使直达瓶底,如有专用灌注装置则更HAO。若为初次使用,加时更要缓慢,以免温度骤降而使容器损坏。细胞冻存时常备的材料有:0.25%胰蛋白酶,含10%~20%的血清培养,DMSO(分析纯)或无色新鲜甘油(121°C蒸气GAO压消毒),2mL安瓿(或专用细胞冻存管)、吸管、离心管、喷灯、纱布袋(或冻存管架)等。主要操作步骤为:(1)选择处于对数生长期的细胞,在冻存前一天ZuiHAO换。将多个培养瓶中的细胞培养 去掉,用0.25%胰蛋白酶消化。适时去掉胰蛋白酶,加入少量新培养。用吸管吸取培养反复吹打瓶壁上的细胞,使其成为均匀分散的细胞悬。悬浮生产细胞则不要消化处理。然后将细胞收集于离心管中离心(1000r/min,10分钟)。(2)去上清,加入含20%小牛血清的完全培养基,于4℃预冷15分钟后,逐滴加入已无菌的DMSO或甘油,用吸管轻轻吹打使细胞均匀,细胞浓度为3×106~1×107/mL之间。(3)将上述细胞分装于安瓿或专用冷冻塑料管中,安瓿装1~1.5mL在火焰喷灯上封口,封口处要完全封闭,圆滑无勾。冷冻管要将盖子盖紧,并标记HAO细胞名称和冻存日期,同时作HAO登记(日期、细胞种类及代次、冻存支数)。(4)将装HAO细胞的安瓿或冻存管装入沙布袋内;置于容器颈口处存放过夜,次日转入中。采用控制降温速度的方法也可采用下列步骤:先将安瓿置入4℃冰箱中2~3小时,再移至冰箱冷冻室内3~4小时,再吊入容器颈气态部分存放2小时,Zui后沉入中。细胞冻存在中可以长期保存,但为妥善起见,冻存半年后,ZuiHAO取出一只安瓿细胞复苏培养,观察生长情况,然后再继续冻存。
TJ905 Cells;背景说明:胶质瘤;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CCC-HEL-1 Cells、SNU-407 Cells、Pa017C Cells
C1498 Cells;背景说明:白血病;雌性;C57BL/6J;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:EB-3 Cells、BJA-B1 Cells、Mono Mac 1 Cells
Daoy Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:6传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:多边形;相关产品有:KMB-17 Cells、UMRC-2 Cells、Rat-2 Cells
L6 Cells;背景说明:该细胞是Yaffe在甲基胆蒽存在的情况下从大鼠大腿肌原代培养的最初两代细胞中分离得到的;在培养基中融合形成多核的肌管和横纹肌纤维,细胞融合的程度随着代数的增加而下降,因此细胞应低代次冷冻并周期性地重新克隆以选择融合能力强的细胞。鼠痘病毒阴性。该细胞应在达汇合状态前传代,以延缓细胞分化能力的丧失。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成肌细胞样;相关产品有:SKCO 1 Cells、DU4475 Cells、Michigan Cancer Foundation-12F Cells
GM03320D Cells;背景说明:该细胞是1967年4月由NicholsWW,LeeJ和DwightS建立,来源于一名13月龄白人男婴腹部肿块,临床诊断为神经母细胞瘤,伴有极少部位的类器官样分化。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:存在两种细胞类型,小的神经母细胞样细胞和大的透明成纤维样细胞;相关产品有:TK.10 Cells、HLEB3 Cells、RH35 Cells
AX-Mel Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:6传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:TCMK-1 Cells、OCUM-1 Cells、AE 1201 Cells
OCI Ly1 Cells;背景说明:弥漫大B淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCIH1437 Cells、P3X63 Ag8.653 Cells、SUDHL-2 Cells
CMT.64 Cells;背景说明:肺腺癌;雌性;C57;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Mv.1.Lu Cells、NCI-H2135 Cells、Mel-RM Cells
K562/ADP Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SJRH-30 Cells、TE-85 clone 5 Cells、Renal Proximal Tubule Epithelial Cells/TERT-immortalized 1 Cells
NCl-H157 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:BC-027 Cells、HITT15 Cells、MDA-134 Cells
H1694 Cells;背景说明:详见相关文献介绍;传代方法:3-4天换液1次。;生长特性:悬浮生长;形态特性:聚团悬浮;相关产品有:beta TC6 Cells、TE671 Cells、CCRF/CEM/0 Cells
HS-294-T Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:混合星状和多边形;相关产品有:HSAEC1-KT Cells、TC-1[JHU-1] Cells、TALL1 Cells
SuDHL 2 Cells;背景说明:弥漫性大细胞淋巴瘤;胸腔积液转移;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:DH 82 Cells、Ly7 Cells、H378 Cells
SK-N-MC Cells;背景说明:这株细胞与HTB-11都是神经源的。1971年9月分离得到SK-N-MC后,发现它有中性多巴胺-β-羟化酶活性,也有细胞内儿茶胺,用甲醛可以诱导出荧光。;传代方法:1:6-1:12传代,每周2-3次换液。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Panc-02 Cells、TE354T Cells、MDA-MB231 Cells
OE-33 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:MEC1 Cells、Hs 934.T Cells、KM12 SM Cells
RMG-I Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:HMCB Cells、C3H-10T1/2 Cells、293T/17 Cells
1E8-H Cells;背景说明:前列腺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:RPMI 7666 Cells、RBSMC Cells、LNCaP Cells
2RS Cells(拥有STR基因鉴定图谱)
Abcam HeLa SPR KO Cells(拥有STR基因鉴定图谱)
Agnes Cells(拥有STR基因鉴定图谱)
BayGenomics ES cell line RRI487 Cells(拥有STR基因鉴定图谱)
BayGenomics ES cell line XI456 Cells(拥有STR基因鉴定图谱)
C2169 Cells(拥有STR基因鉴定图谱)
DA00170 Cells(拥有STR基因鉴定图谱)
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
DA04906 Cells(拥有STR基因鉴定图谱)
FUi002-A Cells(拥有STR基因鉴定图谱)
HEL9217 Cells;背景说明:详见相关文献介绍;传代方法:每周2-3次。;生长特性:悬浮生长;形态特性:成淋巴细胞;相关产品有:Kato-III Cells、H1836 Cells、TE 85.T Cells
OVCA-433 Cells;背景说明:卵巢癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HOS Cells、NCC-IT Cells、Bowes melanoma cells Cells
SUIT 2 Cells;背景说明:胰腺管癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:K562/ADR Cells、P-3J Cells、SUDHL5 Cells
HCT 116人结肠癌传代细胞|送STR图谱
HCC366 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:L-363 Cells、KYSE70 Cells、GM00637H Cells
COLO679 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:EPC Cells、Hs-940 Cells、SCH Cells
TR 146 Cells;背景说明:食管鳞癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SW1222 Cells、SNB19 Cells、KM H-2 Cells
JEG-3 Cells;背景说明:这是一株超三倍体人类细胞株;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:H-295R Cells、GM02131A Cells、Shadyside Hospital Pittsburgh-77 Cells
CHP 100 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:PANC3.27 Cells、RWPE-1 Cells、Hs-611-T Cells
GLC-15 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:LLCMK2 Cells、INS-1E Cells、He-La Cells
Ku812F Cells;背景说明:慢性粒细胞白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:159PT Cells、GC-2 Cells、CD-18 Cells
NCI H69 Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:4传代,每周换液2次;生长特性:悬浮生长,聚团;形态特性:聚团悬浮;相关产品有:SU.86.86 Cells、CAL51 Cells、SNU-387 Cells
SW 954 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:A375-S2 Cells、Mel526 Cells、SUIT 2 Cells
Caco-2 BBe Cells;背景说明:详见相关文献介绍;传代方法:1:6—1:10传代,每周换液2次;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:RPMI 6666 Cells、Human Lung Microvascular Endothelial Cell line-5a Cells、Colo-678 Cells
LuCL4 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:CD 18 Cells、BC-025 Cells、Baby Hamster Kidney 21 Cells
Neukoplast Cells;背景说明:NK细胞;淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:HKF Cells、Laboratory of Allergic Diseases 2 Cells、TOV21G Cells
HAP1 ALOX15 (-) 1 Cells(拥有STR基因鉴定图谱)
HAP1 SLC1A4 (-) Cells(拥有STR基因鉴定图谱)
K7M2-WT Cells;背景说明:骨肉瘤;肺转移;雌性;BALB/c;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SW.620 Cells、CMT93 Cells、SCL-I Cells
X63-Ag8.653 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:LU451 Cells、H-196 Cells、SHIN3 Cells
QBC939 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:HRA-19 Cells、TE9 Cells、C518 Cells
FUOV1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:COLO320 DM Cells、MonoMac 6 Cells、MNNGHOS Cells
RPE1 Cells;背景说明:视网膜色素上皮;hTERT永生;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:WM 266-4 Cells、H-2195 Cells、CCD-112 CoN Cells
3396 Cells;背景说明:乳腺癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MDA MB 361 Cells、Panc 05.04 Cells、PC-3M-2B4 Cells
BN-CL2 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:SW-1271 Cells、OCI/AML-4 Cells、Calu-3 Cells
GM2219C Cells;背景说明:MOLT-4与MOLT-3来源于一名19岁的男性急性淋巴细胞性白血病的复发患者,该患者前期接受过多种药物联合化疗。MOLT-4细胞系为T淋巴细胞起源,p53基因的第248位密码子有一个G→A突变,不表达p53,不表达免疫球蛋白或EB病毒;可产生高水平的末端脱氧核糖转移酶;表达CD1(49%),CD2(35%),CD3A(26%)B(33%)C(34%),CD4(55%),CD5(72%),CD6(22%),CD7(77%)。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:淋巴母细胞样;圆形;相关产品有:AK Cells、HEK 293-EBNA Cells、Loucy Cells
HyCyte THP-1 KO-hIRAK4 Cells(拥有STR基因鉴定图谱)
Le Mor Cells(拥有STR基因鉴定图谱)
NALM6/Clo Cells(拥有STR基因鉴定图谱)
P3X63Ag8.653-Turbodoma Cells(拥有STR基因鉴定图谱)
RUCH-3 Cells(拥有STR基因鉴定图谱)
Ubigene HEK293 ITGA4 KO Cells(拥有STR基因鉴定图谱)
WG1999 Cells(拥有STR基因鉴定图谱)
HEK-SLC15A4-KO-c4 Cells(拥有STR基因鉴定图谱)
U20-S Cells;背景说明:骨肉瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MCF-12F Cells、OPM-2 Cells、HuT-102 Cells
Hs 636.T Cells;背景说明:宫颈鳞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:EFO-27 Cells、HCC1428 Cells、PAa Cells
LA-4 [Mouse lung adenoma] Cells;背景说明:肺癌;A/He;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H2347 Cells、DSL-6A-C1 Cells、RIMEC Cells
IOSE80UBC Cells;背景说明:卵巢;上皮细胞;SV40转化;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HBL1 Cells、SW 962 Cells、NCIH2141 Cells
LC-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H1836 Cells、JB6 clone 30, subclone 7b Cells、CO115 Cells
LC-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H1836 Cells、JB6 clone 30, subclone 7b Cells、CO115 Cells
LA-N-6(OAN) Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Mel RM Cells、MHCC97H Cells、NG 108-15 Cells
SMA-560 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MD Anderson-Metastatic Breast-361 Cells、801-D Cells、Tu-686 Cells
CEK Cells;背景说明:胚肾;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:BEAS 2B Cells、HS 445T Cells、Calu-1 Cells
HEYA8 Cells;背景说明:卵巢癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs675T Cells、SJRH30 Cells、FET Cells
LA-N-5 Cells;背景说明:神经母细胞瘤;骨髓转移;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Merwin Plasma Cell tumor-11 Cells、NIH:OVCAR-8 Cells、LAD-2 Cells
LC-1Sq Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCIH2172 Cells、Hs 281.T Cells、Ramos G6.C10 Cells
MARC145 Cells;背景说明:胚肾;自发永生;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:FRH-0201 Cells、IPLB-SF-21-AE Cells、HCC2157 Cells
SW-1088 Cells;背景说明:星形胶质瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CEMO-1 Cells、C17 Cells、PL45 Cells
SW-579 Cells;背景说明:在裸鼠中成瘤(产生三级恶性纺锤状巨细胞瘤)。 ;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:OCI/AML3 Cells、M-G63 Cells、MAVER Cells
TgSVB Cells(拥有STR基因鉴定图谱)
SMMC-7721 Cells;背景说明:用Northernblot方法,未能检测到细胞中1.3kbLFIRE-1/HFREP-1mRNA的表达。;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:CCD841CoN Cells、NCIH1651 Cells、L Wnt-3A Cells
ACCM Cells;背景说明:涎腺腺样囊性癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Rat Skin 1 Cells、143B Cells、SW626 Cells
MDA-453 Cells;背景说明:该细胞系由CailleauR在1976年从一名48岁的患有转移性乳腺癌的白人女性的心包渗出液中分离建立的。该细胞表达FGF的受体。;传代方法:1:2-1:4传代;2-3天换液1次;生长特性:贴壁生长;形态特性:上皮样;多角形;相关产品有:JCA1 Cells、CHP-100 Cells、Caco-2/BBe 1 Cells
VSMC Cells;背景说明:胸主动脉平滑肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hce8693 Cells、Calu3 Cells、IGROV 1 Cells
B95.8 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:BC-3-H-I Cells、HT29 Cells、Kit-225-K6 Cells
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
UCLA-SO-M21 Cells;背景说明:黑色素瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NK92MI Cells、STO Cells、MV-3 Cells
HLEB3 Cells;背景说明:晶状体;Ad12-SV40转化;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:McA-RH 7777 Cells、2780CP Cells、NRK-52E Cells
SK MEL-28 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:8传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:星形的;相关产品有:Mv 1 Lu Cells、SK-ChA1 Cells、RGCs Cells
KMB17 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:Verda reno Cells、CNE-2Z Cells、801-D Cells
HCT 116人结肠癌传代细胞|送STR图谱
Hs-675-T Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代,每周2-3次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:HO8910 Cells、HSC 3 Cells、RS 4;11 Cells
NCIH520 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;2-3天换液1次;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SUDHL-16 Cells、WEHI 164 TC Cells、TC-1[JHU-1] Cells
SKLMS-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:5传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:NTERA2D1 Cells、LNT-229 Cells、HGMC Cells
H69/P Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:4传代,每周换液2次;生长特性:悬浮生长,聚团;形态特性:聚团悬浮;相关产品有:hSCC-25 Cells、786-0 Cells、MDA PCa 2b Cells
SK N SH Cells;背景说明:SK-N-SH细胞系由J.L.Bieder建系,它与SK-N-MC所不同的是倍增时间较长且多巴胺-β-羟基酶水平较高。 SK-N-SH在细胞介导的细胞毒性试验中用作靶细胞系。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:上皮细胞样;相关产品有:Tu 177 Cells、CP70 Cells、Evsa-T Cells
BayGenomics ES cell line CSH856 Cells(拥有STR基因鉴定图谱)
BayGenomics ES cell line RST122 Cells(拥有STR基因鉴定图谱)
BrMi-J2imm Tlr3-/- Cells(拥有STR基因鉴定图谱)
L1210/6TG Cells(拥有STR基因鉴定图谱)
PPMX0501 Cells(拥有STR基因鉴定图谱)
Rama 29 Cells(拥有STR基因鉴定图谱)
"
传代方法:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
换液频率:每周2-3次
背景资料:是一种人结肠癌细胞系,是由M·Brattain等人于1979年从患结肠癌的男性病人中分离的三株恶性细胞中的一株。HCT 116细胞在半固体琼脂糖培养基中形成克隆;HCT 116细胞在无胸腺裸鼠有致瘤性,形成肿瘤结节。
常见细胞贴壁较弱原因:在遇到运输低温及震荡、室温静置太长时间、添加的培养基或其他试剂过冷、密度较高、聚集未吹散、加液吹打到细胞面等情况时会出现明显的成片脱落现象,此时若脱落现象不严重应尽快放回培养基继续培养,若呈大片脱落的情况时需要收集细胞重新消化吹散并接种;建议使用经过包被或者高贴壁培养瓶培养细胞,尽量避免接触低温或密度过高。
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
VP303 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Huh-7 Cells、COLO 205 Cells、MOLP-8 Cells
BT-20 Cells;背景说明:该细胞1958年由E.Y. Lasfargues 和 L. Ozzello 建系,源自一位74岁白人女性的乳腺癌组织。该细胞表达WNT3和WNT78。TNF alpha抑制该细胞生长。该细胞雌激素受体阴性,但表达5'外显子缺失的雌激素mRNA。;传代方法:1:2—1:4传代,2—3天换液一次;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:Ramos (RA 1) Cells、H-1694 Cells、MC-4 Cells
Hx-147 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:PLB 985 Cells、MES-23.5 Cells、PG-4 Cells
133B Cells(拥有STR基因鉴定图谱)
HCT 116人结肠癌传代细胞|送STR图谱
产品包装形式:复苏细胞:T25培养瓶(一瓶)或冻存细胞:1ml冻存管(两支)
来源说明:细胞主要来源ATCC、DSMZ等细胞库
ATCC细胞库(American Type Culture Colection),该中心一直致力于细胞分类、鉴定和保藏工作。ATCC是全球最大的生物资源保藏中心,ATCC通过行业标准产品、服务和创新解决方案支持全球学术、政府、生物技术、制药、食品、农业和工业领域的科学进步。ATCC提供的服务和定制解决方案包括细胞和微生物培养、鉴定、生物衍生物的开发和生产、性能测试和生物资源保藏服务。美国国家标准协会(ANSI)认可了ATCC标准开发组织,并制定了标准协议,以确保生物材料的可靠性和可重复性。ATCC的使命是为了获取、鉴定、保存、开发、标准化和分发生物资源和生物信息,以提高和应用生物科学知识。
物种来源:Human\Mouse\Rat\Others
HCT 116人结肠癌传代细胞|送STR图谱
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
SNUC2B Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:PaTu 8988 S Cells、H-727 Cells、C-28/I2 Cells
SCC-25 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:10传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:ZR75-1 Cells、HUT 28 Cells、RPMI #1846 Cells
D 407 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:HCT 15 Cells、293 HEK Cells、ECC12 Cells
形态特性:上皮细胞样
细胞冻存复苏材料与方法步骤:常用的细胞冷冻贮存器为贮存器,规格有35L和50L两种。使用时要注意以下几点:(1)一般两周需充一次,至少一个月充一次。温度达-196℃,使用时注意勿让溅到皮肤上,以免引起冻伤。(2)容器为双层结构,中间为真空层,瓶口有双层焊接处,应防止焊接部裂开。(3)在装入时,要注意缓慢小心,并用厚纸卷筒或制漏斗作引导,使直达瓶底,如有专用灌注装置则更HAO。若为初次使用,加时更要缓慢,以免温度骤降而使容器损坏。细胞冻存时常备的材料有:0.25%胰蛋白酶,含10%~20%的血清培养,DMSO(分析纯)或无色新鲜甘油(121°C蒸气GAO压消毒),2mL安瓿(或专用细胞冻存管)、吸管、离心管、喷灯、纱布袋(或冻存管架)等。主要操作步骤为:(1)选择处于对数生长期的细胞,在冻存前一天ZuiHAO换。将多个培养瓶中的细胞培养 去掉,用0.25%胰蛋白酶消化。适时去掉胰蛋白酶,加入少量新培养。用吸管吸取培养反复吹打瓶壁上的细胞,使其成为均匀分散的细胞悬。悬浮生产细胞则不要消化处理。然后将细胞收集于离心管中离心(1000r/min,10分钟)。(2)去上清,加入含20%小牛血清的完全培养基,于4℃预冷15分钟后,逐滴加入已无菌的DMSO或甘油,用吸管轻轻吹打使细胞均匀,细胞浓度为3×106~1×107/mL之间。(3)将上述细胞分装于安瓿或专用冷冻塑料管中,安瓿装1~1.5mL在火焰喷灯上封口,封口处要完全封闭,圆滑无勾。冷冻管要将盖子盖紧,并标记HAO细胞名称和冻存日期,同时作HAO登记(日期、细胞种类及代次、冻存支数)。(4)将装HAO细胞的安瓿或冻存管装入沙布袋内;置于容器颈口处存放过夜,次日转入中。采用控制降温速度的方法也可采用下列步骤:先将安瓿置入4℃冰箱中2~3小时,再移至冰箱冷冻室内3~4小时,再吊入容器颈气态部分存放2小时,Zui后沉入中。细胞冻存在中可以长期保存,但为妥善起见,冻存半年后,ZuiHAO取出一只安瓿细胞复苏培养,观察生长情况,然后再继续冻存。
TJ905 Cells;背景说明:胶质瘤;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CCC-HEL-1 Cells、SNU-407 Cells、Pa017C Cells
C1498 Cells;背景说明:白血病;雌性;C57BL/6J;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:EB-3 Cells、BJA-B1 Cells、Mono Mac 1 Cells
Daoy Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:6传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:多边形;相关产品有:KMB-17 Cells、UMRC-2 Cells、Rat-2 Cells
L6 Cells;背景说明:该细胞是Yaffe在甲基胆蒽存在的情况下从大鼠大腿肌原代培养的最初两代细胞中分离得到的;在培养基中融合形成多核的肌管和横纹肌纤维,细胞融合的程度随着代数的增加而下降,因此细胞应低代次冷冻并周期性地重新克隆以选择融合能力强的细胞。鼠痘病毒阴性。该细胞应在达汇合状态前传代,以延缓细胞分化能力的丧失。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成肌细胞样;相关产品有:SKCO 1 Cells、DU4475 Cells、Michigan Cancer Foundation-12F Cells
GM03320D Cells;背景说明:该细胞是1967年4月由NicholsWW,LeeJ和DwightS建立,来源于一名13月龄白人男婴腹部肿块,临床诊断为神经母细胞瘤,伴有极少部位的类器官样分化。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:存在两种细胞类型,小的神经母细胞样细胞和大的透明成纤维样细胞;相关产品有:TK.10 Cells、HLEB3 Cells、RH35 Cells
AX-Mel Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:6传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:TCMK-1 Cells、OCUM-1 Cells、AE 1201 Cells
OCI Ly1 Cells;背景说明:弥漫大B淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCIH1437 Cells、P3X63 Ag8.653 Cells、SUDHL-2 Cells
CMT.64 Cells;背景说明:肺腺癌;雌性;C57;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Mv.1.Lu Cells、NCI-H2135 Cells、Mel-RM Cells
K562/ADP Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SJRH-30 Cells、TE-85 clone 5 Cells、Renal Proximal Tubule Epithelial Cells/TERT-immortalized 1 Cells
NCl-H157 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:BC-027 Cells、HITT15 Cells、MDA-134 Cells
H1694 Cells;背景说明:详见相关文献介绍;传代方法:3-4天换液1次。;生长特性:悬浮生长;形态特性:聚团悬浮;相关产品有:beta TC6 Cells、TE671 Cells、CCRF/CEM/0 Cells
HS-294-T Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:混合星状和多边形;相关产品有:HSAEC1-KT Cells、TC-1[JHU-1] Cells、TALL1 Cells
SuDHL 2 Cells;背景说明:弥漫性大细胞淋巴瘤;胸腔积液转移;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:DH 82 Cells、Ly7 Cells、H378 Cells
SK-N-MC Cells;背景说明:这株细胞与HTB-11都是神经源的。1971年9月分离得到SK-N-MC后,发现它有中性多巴胺-β-羟化酶活性,也有细胞内儿茶胺,用甲醛可以诱导出荧光。;传代方法:1:6-1:12传代,每周2-3次换液。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Panc-02 Cells、TE354T Cells、MDA-MB231 Cells
OE-33 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:MEC1 Cells、Hs 934.T Cells、KM12 SM Cells
RMG-I Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:HMCB Cells、C3H-10T1/2 Cells、293T/17 Cells
1E8-H Cells;背景说明:前列腺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:RPMI 7666 Cells、RBSMC Cells、LNCaP Cells
2RS Cells(拥有STR基因鉴定图谱)
Abcam HeLa SPR KO Cells(拥有STR基因鉴定图谱)
Agnes Cells(拥有STR基因鉴定图谱)
BayGenomics ES cell line RRI487 Cells(拥有STR基因鉴定图谱)
BayGenomics ES cell line XI456 Cells(拥有STR基因鉴定图谱)
C2169 Cells(拥有STR基因鉴定图谱)
DA00170 Cells(拥有STR基因鉴定图谱)
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
DA04906 Cells(拥有STR基因鉴定图谱)
FUi002-A Cells(拥有STR基因鉴定图谱)
HEL9217 Cells;背景说明:详见相关文献介绍;传代方法:每周2-3次。;生长特性:悬浮生长;形态特性:成淋巴细胞;相关产品有:Kato-III Cells、H1836 Cells、TE 85.T Cells
OVCA-433 Cells;背景说明:卵巢癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HOS Cells、NCC-IT Cells、Bowes melanoma cells Cells
SUIT 2 Cells;背景说明:胰腺管癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:K562/ADR Cells、P-3J Cells、SUDHL5 Cells
HCT 116人结肠癌传代细胞|送STR图谱
HCC366 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:L-363 Cells、KYSE70 Cells、GM00637H Cells
COLO679 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:EPC Cells、Hs-940 Cells、SCH Cells
TR 146 Cells;背景说明:食管鳞癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SW1222 Cells、SNB19 Cells、KM H-2 Cells
JEG-3 Cells;背景说明:这是一株超三倍体人类细胞株;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:H-295R Cells、GM02131A Cells、Shadyside Hospital Pittsburgh-77 Cells
CHP 100 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:PANC3.27 Cells、RWPE-1 Cells、Hs-611-T Cells
GLC-15 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:LLCMK2 Cells、INS-1E Cells、He-La Cells
Ku812F Cells;背景说明:慢性粒细胞白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:159PT Cells、GC-2 Cells、CD-18 Cells
NCI H69 Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:4传代,每周换液2次;生长特性:悬浮生长,聚团;形态特性:聚团悬浮;相关产品有:SU.86.86 Cells、CAL51 Cells、SNU-387 Cells
SW 954 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:A375-S2 Cells、Mel526 Cells、SUIT 2 Cells
Caco-2 BBe Cells;背景说明:详见相关文献介绍;传代方法:1:6—1:10传代,每周换液2次;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:RPMI 6666 Cells、Human Lung Microvascular Endothelial Cell line-5a Cells、Colo-678 Cells
LuCL4 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:CD 18 Cells、BC-025 Cells、Baby Hamster Kidney 21 Cells
Neukoplast Cells;背景说明:NK细胞;淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:HKF Cells、Laboratory of Allergic Diseases 2 Cells、TOV21G Cells
HAP1 ALOX15 (-) 1 Cells(拥有STR基因鉴定图谱)
HAP1 SLC1A4 (-) Cells(拥有STR基因鉴定图谱)
K7M2-WT Cells;背景说明:骨肉瘤;肺转移;雌性;BALB/c;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SW.620 Cells、CMT93 Cells、SCL-I Cells
X63-Ag8.653 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:LU451 Cells、H-196 Cells、SHIN3 Cells
QBC939 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:HRA-19 Cells、TE9 Cells、C518 Cells
FUOV1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:COLO320 DM Cells、MonoMac 6 Cells、MNNGHOS Cells
RPE1 Cells;背景说明:视网膜色素上皮;hTERT永生;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:WM 266-4 Cells、H-2195 Cells、CCD-112 CoN Cells
3396 Cells;背景说明:乳腺癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MDA MB 361 Cells、Panc 05.04 Cells、PC-3M-2B4 Cells
BN-CL2 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:SW-1271 Cells、OCI/AML-4 Cells、Calu-3 Cells
GM2219C Cells;背景说明:MOLT-4与MOLT-3来源于一名19岁的男性急性淋巴细胞性白血病的复发患者,该患者前期接受过多种药物联合化疗。MOLT-4细胞系为T淋巴细胞起源,p53基因的第248位密码子有一个G→A突变,不表达p53,不表达免疫球蛋白或EB病毒;可产生高水平的末端脱氧核糖转移酶;表达CD1(49%),CD2(35%),CD3A(26%)B(33%)C(34%),CD4(55%),CD5(72%),CD6(22%),CD7(77%)。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:淋巴母细胞样;圆形;相关产品有:AK Cells、HEK 293-EBNA Cells、Loucy Cells
HyCyte THP-1 KO-hIRAK4 Cells(拥有STR基因鉴定图谱)
Le Mor Cells(拥有STR基因鉴定图谱)
NALM6/Clo Cells(拥有STR基因鉴定图谱)
P3X63Ag8.653-Turbodoma Cells(拥有STR基因鉴定图谱)
RUCH-3 Cells(拥有STR基因鉴定图谱)
Ubigene HEK293 ITGA4 KO Cells(拥有STR基因鉴定图谱)
WG1999 Cells(拥有STR基因鉴定图谱)
HEK-SLC15A4-KO-c4 Cells(拥有STR基因鉴定图谱)
U20-S Cells;背景说明:骨肉瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MCF-12F Cells、OPM-2 Cells、HuT-102 Cells
Hs 636.T Cells;背景说明:宫颈鳞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:EFO-27 Cells、HCC1428 Cells、PAa Cells
LA-4 [Mouse lung adenoma] Cells;背景说明:肺癌;A/He;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H2347 Cells、DSL-6A-C1 Cells、RIMEC Cells
IOSE80UBC Cells;背景说明:卵巢;上皮细胞;SV40转化;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HBL1 Cells、SW 962 Cells、NCIH2141 Cells
LC-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H1836 Cells、JB6 clone 30, subclone 7b Cells、CO115 Cells
LC-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H1836 Cells、JB6 clone 30, subclone 7b Cells、CO115 Cells
LA-N-6(OAN) Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Mel RM Cells、MHCC97H Cells、NG 108-15 Cells
SMA-560 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MD Anderson-Metastatic Breast-361 Cells、801-D Cells、Tu-686 Cells
CEK Cells;背景说明:胚肾;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:BEAS 2B Cells、HS 445T Cells、Calu-1 Cells
HEYA8 Cells;背景说明:卵巢癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs675T Cells、SJRH30 Cells、FET Cells
LA-N-5 Cells;背景说明:神经母细胞瘤;骨髓转移;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Merwin Plasma Cell tumor-11 Cells、NIH:OVCAR-8 Cells、LAD-2 Cells
LC-1Sq Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCIH2172 Cells、Hs 281.T Cells、Ramos G6.C10 Cells
MARC145 Cells;背景说明:胚肾;自发永生;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:FRH-0201 Cells、IPLB-SF-21-AE Cells、HCC2157 Cells
SW-1088 Cells;背景说明:星形胶质瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CEMO-1 Cells、C17 Cells、PL45 Cells
SW-579 Cells;背景说明:在裸鼠中成瘤(产生三级恶性纺锤状巨细胞瘤)。 ;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:OCI/AML3 Cells、M-G63 Cells、MAVER Cells
TgSVB Cells(拥有STR基因鉴定图谱)
SMMC-7721 Cells;背景说明:用Northernblot方法,未能检测到细胞中1.3kbLFIRE-1/HFREP-1mRNA的表达。;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:CCD841CoN Cells、NCIH1651 Cells、L Wnt-3A Cells
ACCM Cells;背景说明:涎腺腺样囊性癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Rat Skin 1 Cells、143B Cells、SW626 Cells
MDA-453 Cells;背景说明:该细胞系由CailleauR在1976年从一名48岁的患有转移性乳腺癌的白人女性的心包渗出液中分离建立的。该细胞表达FGF的受体。;传代方法:1:2-1:4传代;2-3天换液1次;生长特性:贴壁生长;形态特性:上皮样;多角形;相关产品有:JCA1 Cells、CHP-100 Cells、Caco-2/BBe 1 Cells
VSMC Cells;背景说明:胸主动脉平滑肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hce8693 Cells、Calu3 Cells、IGROV 1 Cells
B95.8 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:BC-3-H-I Cells、HT29 Cells、Kit-225-K6 Cells
┈订┈购┈热┈线:1┈5┈8┈0┈0┈5┈7┈6┈8┈6┈7【微信同号】┈Q┈Q:3┈3┈0┈7┈2┈0┈4┈2┈7┈1;
UCLA-SO-M21 Cells;背景说明:黑色素瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NK92MI Cells、STO Cells、MV-3 Cells
HLEB3 Cells;背景说明:晶状体;Ad12-SV40转化;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:McA-RH 7777 Cells、2780CP Cells、NRK-52E Cells
SK MEL-28 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:8传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:星形的;相关产品有:Mv 1 Lu Cells、SK-ChA1 Cells、RGCs Cells
KMB17 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:Verda reno Cells、CNE-2Z Cells、801-D Cells
HCT 116人结肠癌传代细胞|送STR图谱
Hs-675-T Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代,每周2-3次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:HO8910 Cells、HSC 3 Cells、RS 4;11 Cells
NCIH520 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;2-3天换液1次;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SUDHL-16 Cells、WEHI 164 TC Cells、TC-1[JHU-1] Cells
SKLMS-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:5传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:NTERA2D1 Cells、LNT-229 Cells、HGMC Cells
H69/P Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:4传代,每周换液2次;生长特性:悬浮生长,聚团;形态特性:聚团悬浮;相关产品有:hSCC-25 Cells、786-0 Cells、MDA PCa 2b Cells
SK N SH Cells;背景说明:SK-N-SH细胞系由J.L.Bieder建系,它与SK-N-MC所不同的是倍增时间较长且多巴胺-β-羟基酶水平较高。 SK-N-SH在细胞介导的细胞毒性试验中用作靶细胞系。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:上皮细胞样;相关产品有:Tu 177 Cells、CP70 Cells、Evsa-T Cells
BayGenomics ES cell line CSH856 Cells(拥有STR基因鉴定图谱)
BayGenomics ES cell line RST122 Cells(拥有STR基因鉴定图谱)
BrMi-J2imm Tlr3-/- Cells(拥有STR基因鉴定图谱)
L1210/6TG Cells(拥有STR基因鉴定图谱)
PPMX0501 Cells(拥有STR基因鉴定图谱)
Rama 29 Cells(拥有STR基因鉴定图谱)
"
风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验该产品被引用文献
"PubMed=2835152
Boyd D., Florent G., Kim P., Brattain M.G.
Determination of the levels of urokinase and its receptor in human colon carcinoma cell lines.
Cancer Res. 48:3112-3116(1988)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045; PMCID=PMC45163
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=7761852; DOI=10.1126/science.7761852
Markowitz S.D., Wang J., Myeroff L.L., Parsons R., Sun L.-Z., Lutterbaugh J.D., Fan R.S., Zborowska E., Kinzler K.W., Vogelstein B., Brattain M.G., Willson J.K.V.
Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability.
Science 268:1336-1338(1995)
PubMed=7824277
Eshleman J.R., Lang E.Z., Bowerfind G.K., Parsons R., Vogelstein B., Willson J.K.V., Veigl M.L., Sedwick W.D., Markowitz S.D.
Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer.
Oncogene 10:33-37(1995)
PubMed=9000147
Cottu P.-H., Muzeau F., Estreicher A., Flejou J.-F., Iggo R.D., Thomas G., Hamelin R.
Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines.
Oncogene 13:2727-2730(1996)
PubMed=9000572
Hoang J.-M., Cottu P.-H., Thuille B., Salmon R.J., Thomas G., Hamelin R.
BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines.
Cancer Res. 57:300-303(1997)
PubMed=9023415; DOI=10.1006/cimm.1996.1062
Seki N., Hoshino T., Kikuchi M., Hayashi A., Itoh K.
HLA-A locus-restricted and tumor-specific CTLs in tumor-infiltrating lymphocytes of patients with non-small cell lung cancer.
Cell. Immunol. 175:101-110(1997)
PubMed=9178645; DOI=10.1006/cimm.1997.1108
Nakao M., Sata M., Saitsu H., Yutani S., Kawamoto M., Kojiro M., Itoh K.
CD4+ hepatic cancer-specific cytotoxic T lymphocytes in patients with hepatocellular carcinoma.
Cell. Immunol. 177:176-181(1997)
PubMed=9294210; DOI=10.1073/pnas.94.19.10330; PMCID=PMC23362
Ilyas M., Tomlinson I.P.M., Rowan A.J., Pignatelli M., Bodmer W.F.
Beta-catenin mutations in cell lines established from human colorectal cancers.
Proc. Natl. Acad. Sci. U.S.A. 94:10330-10334(1997)
PubMed=9515795
Sparks A.B., Morin P.J., Vogelstein B., Kinzler K.W.
Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer.
Cancer Res. 58:1130-1134(1998)
PubMed=9715273; DOI=10.1038/sj.onc.1201986
Eshleman J.R., Casey G., Kochera M.E., Sedwick W.D., Swinler S.E., Veigl M.L., Willson J.K.V., Schwartz S., Markowitz S.D.
Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53.
Oncogene 17:719-725(1998)
PubMed=10674020; DOI=10.1016/S0959-8049(99)00206-3
Ku J.-L., Yoon K.-A., Kim D.-Y., Park J.-G.
Mutations in hMSH6 alone are not sufficient to cause the microsatellite instability in colorectal cancer cell lines.
Eur. J. Cancer 35:1724-1729(1999)
PubMed=10612807; DOI=10.1002/(SICI)1098-2264(200002)27:2<183::AID-GCC10>3.0.CO;2-P; PMCID=PMC4721570
Ghadimi B.M., Sackett D.L., Difilippantonio M.J., Schrock E., Neumann T., Jauho A., Auer G., Ried T.
Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations.
Genes Chromosomes Cancer 27:183-190(2000)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10700188; DOI=10.1038/73536
Gayther S.A., Batley S.J., Linger L., Bannister A.J., Thorpe K., Chin S.-F., Daigo Y., Russell P., Wilson A., Sowter H.M., Delhanty J.D.A., Ponder B.A.J., Kouzarides T., Caldas C.
Mutations truncating the EP300 acetylase in human cancers.
Nat. Genet. 24:300-303(2000)
PubMed=10737795; DOI=10.1073/pnas.97.7.3352; PMCID=PMC16243
Rowan A.J., Lamlum H., Ilyas M., Wheeler J.M.D., Straub J., Papadopoulou A., Bicknell D.C., Bodmer W.F., Tomlinson I.P.M.
APC mutations in sporadic colorectal tumors: a mutational 'hotspot' and interdependence of the 'two hits'.
Proc. Natl. Acad. Sci. U.S.A. 97:3352-3357(2000)
PubMed=11226274; DOI=10.1073/pnas.041603298; PMCID=PMC30173
Abdel-Rahman W.M., Katsura K., Rens W., Gorman P.A., Sheer D., Bicknell D.C., Bodmer W.F., Arends M.J., Wyllie A.H., Edwards P.A.W.
Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement.
Proc. Natl. Acad. Sci. U.S.A. 98:2538-2543(2001)
PubMed=11314036; DOI=10.1038/sj.onc.1204211
Forgacs E., Wren J.D., Kamibayashi C., Kondo M., Xu X.L., Markowitz S.D., Tomlinson G.E., Muller C.Y., Gazdar A.F., Garner H.R., Minna J.D.
Searching for microsatellite mutations in coding regions in lung, breast, ovarian and colorectal cancers.
Oncogene 20:1005-1009(2001)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=11526487; DOI=10.1038/sj.onc.1204611
Gayet J., Zhou X.-P., Duval A., Rolland S., Hoang J.-M., Cottu P.-H., Hamelin R.
Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines.
Oncogene 20:5025-5032(2001)
PubMed=11687795; DOI=10.1038/ng754
Snijders A.M., Nowak N.J., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Law S., Myambo K., Palmer J., Ylstra B., Yue J.P., Gray J.W., Jain A.N., Pinkel D., Albertson D.G.
Assembly of microarrays for genome-wide measurement of DNA copy number.
Nat. Genet. 29:263-264(2001)
PubMed=12068308; DOI=10.1038/nature00766
Davies H.R., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=12584437; DOI=10.1159/000068544
Melcher R., Koehler S., Steinlein C., Schmid M., Mueller C.R., Luehrs H., Menzel T., Scheppach W., Moerk H., Scheurlen M., Koehrle J., Al-Taie O.
Spectral karyotype analysis of colon cancer cell lines of the tumor suppressor and mutator pathway.
Cytogenet. Genome Res. 98:22-28(2002)
PubMed=12615714
Hempen P.M., Zhang L., Bansal R.K., Iacobuzio-Donahue C.A., Murphy K.M., Maitra A., Vogelstein B., Whitehead R.H., Markowitz S.D., Willson J.K.V., Yeo C.J., Hruban R.H., Kern S.E.
Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers.
Cancer Res. 63:994-999(2003)
PubMed=12671075; DOI=10.1073/pnas.0831040100; PMCID=PMC153619
Jongeneel C.V., Iseli C., Stevenson B.J., Riggins G.J., Lal A., Mackay A., Harris R.A., O'Hare M.J., Neville A.M., Simpson A.J.G., Strausberg R.L.
Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing.
Proc. Natl. Acad. Sci. U.S.A. 100:4702-4705(2003)
PubMed=12714694; DOI=10.1093/mutage/18.3.277
Yamada N.A., Castro A., Farber R.A.
Variation in the extent of microsatellite instability in human cell lines with defects in different mismatch repair genes.
Mutagenesis 18:277-282(2003)
PubMed=15748285; DOI=10.1186/1479-5876-3-11; PMCID=PMC555742
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. 3rd, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=15900046; DOI=10.1093/jnci/dji133
Mashima T., Oh-hara T., Sato S., Mochizuki M., Sugimoto Y., Yamazaki K., Hamada J.-i., Tada M., Moriuchi T., Ishikawa Y., Kato Y., Tomoda H., Yamori T., Tsuruo T.
p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target.
J. Natl. Cancer Inst. 97:765-777(2005)
PubMed=16418264; DOI=10.1073/pnas.0510146103; PMCID=PMC1327731
Liu Y., Bodmer W.F.
Analysis of p53 mutations and their expression in 56 colorectal cancer cell lines.
Proc. Natl. Acad. Sci. U.S.A. 103:976-981(2006)
PubMed=16854228; DOI=10.1186/1476-4598-5-29; PMCID=PMC1550420
Bandres Elizalde E.M., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J.
Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues.
Mol. Cancer 5:29.1-29.10(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433; PMCID=PMC2705832
Ikediobi O.N., Davies H.R., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Miroo T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=17178751; DOI=10.1093/nar/gkl1030; PMCID=PMC1807964
Fiegler H., Geigl J.B., Langer S., Rigler D., Porter K., Unger K., Carter N.P., Speicher M.R.
High resolution array-CGH analysis of single cells.
Nucleic Acids Res. 35:e15.1-e15.10(2007)
PubMed=17363507; DOI=10.1158/1535-7163.MCT-06-0555
Wang J., Kuropatwinski K.K., Hauser J., Rossi M.R., Zhou Y.-F., Conway A., Kan J.L.C., Gibson N.W., Willson J.K.V., Cowell J.K., Brattain M.G.
Colon carcinoma cells harboring PIK3CA mutations display resistance to growth factor deprivation induced apoptosis.
Mol. Cancer Ther. 6:1143-1150(2007)
PubMed=18258742; DOI=10.1073/pnas.0712176105; PMCID=PMC2268141
Emaduddin M., Bicknell D.C., Bodmer W.F., Feller S.M.
Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells.
Proc. Natl. Acad. Sci. U.S.A. 105:2358-2362(2008)
PubMed=18340113; DOI=10.4161/cbt.7.6.5838
Gongora C., Candeil L., Vezzio-Vie N., Copois V., Denis V., Bareil C., Molina F., Fraslon C., Conseiller E., Pau B., Martineau P., Del Rio M.
Altered expression of cell proliferation-related and interferon-stimulated genes in colon cancer cells resistant to SN38.
Cancer Biol. Ther. 7:822-832(2008)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921; PMCID=PMC4020356
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=19927377; DOI=10.1002/gcc.20730; PMCID=PMC2818350
Knutsen T., Padilla-Nash H.M., Wangsa D., Barenboim-Stapleton L., Camps J., McNeil N.E., Difilippantonio M.J., Ried T.
Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines.
Genes Chromosomes Cancer 49:204-223(2010)
PubMed=20164919; DOI=10.1038/nature08768; PMCID=PMC3145113
Bignell G.R., Greenman C.D., Davies H.R., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192; PMCID=PMC2943514
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780; PMCID=PMC2920028
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=22068913; DOI=10.1073/pnas.1111840108; PMCID=PMC3219108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=21912889; DOI=10.1007/s10637-011-9744-z
Sutherland H.S., Hwang I.Y., Marshall E.S., Lindsay B.S., Denny W.A., Gilchrist C., Joseph W.R., Greenhalgh D., Richardson E., Kestell P., Ding A., Baguley B.C.
Therapeutic reactivation of mutant p53 protein by quinazoline derivatives.
Invest. New Drugs 30:2035-2045(2012)
PubMed=22336246; DOI=10.1016/j.bmc.2012.01.017
Kong D.-X., Yamori T.
JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs.
Bioorg. Med. Chem. 20:1947-1951(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628; PMCID=PMC3276511
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096; PMCID=PMC3285665
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595; PMCID=PMC3526189
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=23272949; DOI=10.1186/1755-8794-5-66; PMCID=PMC3543849
Schlicker A., Beran G., Chresta C.M., McWalter G., Pritchard A., Weston S., Runswick S., Davenport S., Heathcote K., Castro D.A., Orphanides G., French T., Wessels L.F.A.
Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines.
BMC Med. Genomics 5:66.1-66.15(2012)
PubMed=23546019; DOI=10.3892/ijo.2013.1868
Petitprez A., Poindessous V., Ouaret D., Regairaz M., Bastian G., Guerin E., Escargueil A.E., Larsen A.K.
Acquired irinotecan resistance is accompanied by stable modifications of cell cycle dynamics independent of MSI status.
Int. J. Oncol. 42:1644-1653(2013)
PubMed=23631600; DOI=10.1021/pr400260h
Loftus N.J., Lai L., Wilkinson R.W., Odedra R., Wilson I.D., Barnes A.J.
Global metabolite profiling of human colorectal cancer xenografts in mice using HPLC-MS/MS.
J. Proteome Res. 12:2980-2986(2013)
PubMed=23649806; DOI=10.1083/jcb.201210031; PMCID=PMC3653305
Kleiblova P., Shaltiel I.A., Benada J., Sevcik J., Pechackova S., Pohlreich P., Voest E.E., Dundr P., Bartek J., Kleibl Z., Medema R.H., Macurek L.
Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint.
J. Cell Biol. 201:511-521(2013)
PubMed=23671654; DOI=10.1371/journal.pone.0063056; PMCID=PMC3646030
Lu Y.-H., Soong T.D., Elemento O.
A novel approach for characterizing microsatellite instability in cancer cells.
PLoS ONE 8:E63056-E63056(2013)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342; PMCID=PMC4893961
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24042735; DOI=10.1038/oncsis.2013.35; PMCID=PMC3816225
Ahmed D., Eide P.W., Eilertsen I.A., Danielsen S.A., Eknaes M., Hektoen M., Lind G.E., Lothe R.A.
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis 2:e71.1-e71.8(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20; PMCID=PMC4178206
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24670534; DOI=10.1371/journal.pone.0092047; PMCID=PMC3966786
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893; PMCID=PMC4355981
Boegel S., Lower M., Bukur T., Sahin U., Castle J.C.
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines.
OncoImmunology 3:e954893.1-e954893.12(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35; PMCID=PMC4432652
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25576301; DOI=10.1074/mcp.M114.042812; PMCID=PMC4349985
Bassani-Sternberg M., Pletscher-Frankild S., Jensen L.J., Mann M.
Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation.
Mol. Cell. Proteomics 14:658-673(2015)
PubMed=25841592; DOI=10.1016/j.jprot.2015.03.019
Piersma S.R., Knol J.C., de Reus I., Labots M., Sampadi B.K., Pham T.V., Ishihama Y., Verheul H.M.W., Jimenez C.R.
Feasibility of label-free phosphoproteomics and application to base-line signaling of colorectal cancer cell lines.
J. Proteomics 127:247-258(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=26169745; DOI=10.1186/s12967-015-0576-z; PMCID=PMC4499939
Halama A., Guerrouahen B.S., Pasquier J., Diboun I., Karoly E.D., Suhre K., Rafii A.
Metabolic signatures differentiate ovarian from colon cancer cell lines.
J. Transl. Med. 13:223.1-223.12(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26719794; DOI=10.1186/s13742-015-0106-1; PMCID=PMC4696294
Teo A.S.M., Verzotto D., Yao F., Nagarajan N., Hillmer A.M.
Single-molecule optical genome mapping of a human HapMap and a colorectal cancer cell line.
GigaScience 4:65.1-65.6(2015)
PubMed=26537799; DOI=10.1074/mcp.M115.051235; PMCID=PMC4762531
Holst S., Deuss A.J.M., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A.M., Deelder A.M., Mesker W.E., Tollenaar R.A.E.M., Rombouts Y., Wuhrer M.
N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression.
Mol. Cell. Proteomics 15:124-140(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52; PMCID=PMC4932877
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4; PMCID=PMC5087121
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=28179481; DOI=10.1158/1535-7163.MCT-16-0578
Tanaka N., Mashima T., Mizutani A., Sato A., Aoyama A., Gong B., Yoshida H., Muramatsu Y., Nakata K., Matsuura M., Katayama R., Nagayama S., Fujita N., Sugimoto Y., Seimiya H.
APC mutations as a potential biomarker for sensitivity to tankyrase inhibitors in colorectal cancer.
Mol. Cancer Ther. 16:752-762(2017)
PubMed=28192450; DOI=10.1371/journal.pone.0171435; PMCID=PMC5305277
Fasterius E., Raso C., Kennedy S.A., Rauch N., Lundin P., Kolch W., Uhlen M., Al-Khalili Szigyarto C.
A novel RNA sequencing data analysis method for cell line authentication.
PLoS ONE 12:E0171435-E0171435(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28601559; DOI=10.1016/j.cels.2017.05.009; PMCID=PMC5493283
Bekker-Jensen D.B., Kelstrup C.D., Batth T.S., Larsen S.C., Haldrup C., Bramsen J.B., Sorensen K.D., Hoyer S., Orntoft T.F., Lindbjerg Andersen C., Nielsen M.L., Olsen J.V.
An optimized shotgun strategy for the rapid generation of comprehensive human proteomes.
Cell Syst. 4:587-599.e4(2017)
PubMed=28683746; DOI=10.1186/s12943-017-0691-y; PMCID=PMC5498998
Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A.
Multi-omics of 34 colorectal cancer cell lines -- a resource for biomedical studies.
Mol. Cancer 16:116.1-116.16(2017)
PubMed=28854368; DOI=10.1016/j.celrep.2017.08.010; PMCID=PMC5583477
Roumeliotis T.I., Williams S.P., Goncalves E., Alsinet C., Del Castillo Velasco-Herrera M., Aben N., Ghavidel F.Z., Michaut M., Schubert M., Price S., Wright J.C., Yu L., Yang M., Dienstmann R., Guinney J.H., Beltrao P., Brazma A., Pardo M., Stegle O., Adams D.J., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Choudhary J.S.
Genomic determinants of protein abundance variation in colorectal cancer cells.
Cell Rep. 20:2201-2214(2017)
PubMed=29101300; DOI=10.15252/msb.20177701; PMCID=PMC5731344
Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R.-S., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A., Heinzlmeir S., Johnstone E., Domingo E., Kerr D.J., Jesinghaus M., Slotta-Huspenina J., Weichert W., Knapp S., Feller S.M., Kuster B.
Pharmacoproteomic characterisation of human colon and rectal cancer.
Mol. Syst. Biol. 13:951-951(2017)
PubMed=29207035; DOI=10.3892/ijo.2017.4206
Olejniczak A., Szarynska M., Kmiec Z.
In vitro characterization of spheres derived from colorectal cancer cell lines.
Int. J. Oncol. 52:599-612(2018)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051; PMCID=PMC6343826
Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D.A., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F.
Differential effector engagement by oncogenic KRAS.
Cell Rep. 22:1889-1902(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023; PMCID=PMC7339254
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=32172478; DOI=10.1007/s12253-020-00805-3
Xu Y.-T., Zhang L., Wang Q.-L., Zheng M.-J.
Comparison of different colorectal cancer with liver metastases models using six colorectal cancer cell lines.
Pathol. Oncol. Res. 26:2177-2183(2020)
PubMed=32927768; DOI=10.3390/cancers12092582; PMCID=PMC7564713
Schulte am Esch J., Windmoller B.A., Hanewinkel J., Storm J., Forster C., Wilkens L., Kruger M., Kaltschmidt B., Kaltschmidt C.
Isolation and characterization of two novel colorectal cancer cell lines, containing a subpopulation with potential stem-like properties: treatment options by MYC/NMYC inhibition.
Cancers (Basel) 12:2582.1-2582.34(2020)"
Boyd D., Florent G., Kim P., Brattain M.G.
Determination of the levels of urokinase and its receptor in human colon carcinoma cell lines.
Cancer Res. 48:3112-3116(1988)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045; PMCID=PMC45163
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=7761852; DOI=10.1126/science.7761852
Markowitz S.D., Wang J., Myeroff L.L., Parsons R., Sun L.-Z., Lutterbaugh J.D., Fan R.S., Zborowska E., Kinzler K.W., Vogelstein B., Brattain M.G., Willson J.K.V.
Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability.
Science 268:1336-1338(1995)
PubMed=7824277
Eshleman J.R., Lang E.Z., Bowerfind G.K., Parsons R., Vogelstein B., Willson J.K.V., Veigl M.L., Sedwick W.D., Markowitz S.D.
Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer.
Oncogene 10:33-37(1995)
PubMed=9000147
Cottu P.-H., Muzeau F., Estreicher A., Flejou J.-F., Iggo R.D., Thomas G., Hamelin R.
Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines.
Oncogene 13:2727-2730(1996)
PubMed=9000572
Hoang J.-M., Cottu P.-H., Thuille B., Salmon R.J., Thomas G., Hamelin R.
BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines.
Cancer Res. 57:300-303(1997)
PubMed=9023415; DOI=10.1006/cimm.1996.1062
Seki N., Hoshino T., Kikuchi M., Hayashi A., Itoh K.
HLA-A locus-restricted and tumor-specific CTLs in tumor-infiltrating lymphocytes of patients with non-small cell lung cancer.
Cell. Immunol. 175:101-110(1997)
PubMed=9178645; DOI=10.1006/cimm.1997.1108
Nakao M., Sata M., Saitsu H., Yutani S., Kawamoto M., Kojiro M., Itoh K.
CD4+ hepatic cancer-specific cytotoxic T lymphocytes in patients with hepatocellular carcinoma.
Cell. Immunol. 177:176-181(1997)
PubMed=9294210; DOI=10.1073/pnas.94.19.10330; PMCID=PMC23362
Ilyas M., Tomlinson I.P.M., Rowan A.J., Pignatelli M., Bodmer W.F.
Beta-catenin mutations in cell lines established from human colorectal cancers.
Proc. Natl. Acad. Sci. U.S.A. 94:10330-10334(1997)
PubMed=9515795
Sparks A.B., Morin P.J., Vogelstein B., Kinzler K.W.
Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer.
Cancer Res. 58:1130-1134(1998)
PubMed=9715273; DOI=10.1038/sj.onc.1201986
Eshleman J.R., Casey G., Kochera M.E., Sedwick W.D., Swinler S.E., Veigl M.L., Willson J.K.V., Schwartz S., Markowitz S.D.
Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53.
Oncogene 17:719-725(1998)
PubMed=10674020; DOI=10.1016/S0959-8049(99)00206-3
Ku J.-L., Yoon K.-A., Kim D.-Y., Park J.-G.
Mutations in hMSH6 alone are not sufficient to cause the microsatellite instability in colorectal cancer cell lines.
Eur. J. Cancer 35:1724-1729(1999)
PubMed=10612807; DOI=10.1002/(SICI)1098-2264(200002)27:2<183::AID-GCC10>3.0.CO;2-P; PMCID=PMC4721570
Ghadimi B.M., Sackett D.L., Difilippantonio M.J., Schrock E., Neumann T., Jauho A., Auer G., Ried T.
Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations.
Genes Chromosomes Cancer 27:183-190(2000)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10700188; DOI=10.1038/73536
Gayther S.A., Batley S.J., Linger L., Bannister A.J., Thorpe K., Chin S.-F., Daigo Y., Russell P., Wilson A., Sowter H.M., Delhanty J.D.A., Ponder B.A.J., Kouzarides T., Caldas C.
Mutations truncating the EP300 acetylase in human cancers.
Nat. Genet. 24:300-303(2000)
PubMed=10737795; DOI=10.1073/pnas.97.7.3352; PMCID=PMC16243
Rowan A.J., Lamlum H., Ilyas M., Wheeler J.M.D., Straub J., Papadopoulou A., Bicknell D.C., Bodmer W.F., Tomlinson I.P.M.
APC mutations in sporadic colorectal tumors: a mutational 'hotspot' and interdependence of the 'two hits'.
Proc. Natl. Acad. Sci. U.S.A. 97:3352-3357(2000)
PubMed=11226274; DOI=10.1073/pnas.041603298; PMCID=PMC30173
Abdel-Rahman W.M., Katsura K., Rens W., Gorman P.A., Sheer D., Bicknell D.C., Bodmer W.F., Arends M.J., Wyllie A.H., Edwards P.A.W.
Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement.
Proc. Natl. Acad. Sci. U.S.A. 98:2538-2543(2001)
PubMed=11314036; DOI=10.1038/sj.onc.1204211
Forgacs E., Wren J.D., Kamibayashi C., Kondo M., Xu X.L., Markowitz S.D., Tomlinson G.E., Muller C.Y., Gazdar A.F., Garner H.R., Minna J.D.
Searching for microsatellite mutations in coding regions in lung, breast, ovarian and colorectal cancers.
Oncogene 20:1005-1009(2001)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=11526487; DOI=10.1038/sj.onc.1204611
Gayet J., Zhou X.-P., Duval A., Rolland S., Hoang J.-M., Cottu P.-H., Hamelin R.
Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines.
Oncogene 20:5025-5032(2001)
PubMed=11687795; DOI=10.1038/ng754
Snijders A.M., Nowak N.J., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Law S., Myambo K., Palmer J., Ylstra B., Yue J.P., Gray J.W., Jain A.N., Pinkel D., Albertson D.G.
Assembly of microarrays for genome-wide measurement of DNA copy number.
Nat. Genet. 29:263-264(2001)
PubMed=12068308; DOI=10.1038/nature00766
Davies H.R., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=12584437; DOI=10.1159/000068544
Melcher R., Koehler S., Steinlein C., Schmid M., Mueller C.R., Luehrs H., Menzel T., Scheppach W., Moerk H., Scheurlen M., Koehrle J., Al-Taie O.
Spectral karyotype analysis of colon cancer cell lines of the tumor suppressor and mutator pathway.
Cytogenet. Genome Res. 98:22-28(2002)
PubMed=12615714
Hempen P.M., Zhang L., Bansal R.K., Iacobuzio-Donahue C.A., Murphy K.M., Maitra A., Vogelstein B., Whitehead R.H., Markowitz S.D., Willson J.K.V., Yeo C.J., Hruban R.H., Kern S.E.
Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers.
Cancer Res. 63:994-999(2003)
PubMed=12671075; DOI=10.1073/pnas.0831040100; PMCID=PMC153619
Jongeneel C.V., Iseli C., Stevenson B.J., Riggins G.J., Lal A., Mackay A., Harris R.A., O'Hare M.J., Neville A.M., Simpson A.J.G., Strausberg R.L.
Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing.
Proc. Natl. Acad. Sci. U.S.A. 100:4702-4705(2003)
PubMed=12714694; DOI=10.1093/mutage/18.3.277
Yamada N.A., Castro A., Farber R.A.
Variation in the extent of microsatellite instability in human cell lines with defects in different mismatch repair genes.
Mutagenesis 18:277-282(2003)
PubMed=15748285; DOI=10.1186/1479-5876-3-11; PMCID=PMC555742
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. 3rd, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=15900046; DOI=10.1093/jnci/dji133
Mashima T., Oh-hara T., Sato S., Mochizuki M., Sugimoto Y., Yamazaki K., Hamada J.-i., Tada M., Moriuchi T., Ishikawa Y., Kato Y., Tomoda H., Yamori T., Tsuruo T.
p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target.
J. Natl. Cancer Inst. 97:765-777(2005)
PubMed=16418264; DOI=10.1073/pnas.0510146103; PMCID=PMC1327731
Liu Y., Bodmer W.F.
Analysis of p53 mutations and their expression in 56 colorectal cancer cell lines.
Proc. Natl. Acad. Sci. U.S.A. 103:976-981(2006)
PubMed=16854228; DOI=10.1186/1476-4598-5-29; PMCID=PMC1550420
Bandres Elizalde E.M., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J.
Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues.
Mol. Cancer 5:29.1-29.10(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433; PMCID=PMC2705832
Ikediobi O.N., Davies H.R., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Miroo T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=17178751; DOI=10.1093/nar/gkl1030; PMCID=PMC1807964
Fiegler H., Geigl J.B., Langer S., Rigler D., Porter K., Unger K., Carter N.P., Speicher M.R.
High resolution array-CGH analysis of single cells.
Nucleic Acids Res. 35:e15.1-e15.10(2007)
PubMed=17363507; DOI=10.1158/1535-7163.MCT-06-0555
Wang J., Kuropatwinski K.K., Hauser J., Rossi M.R., Zhou Y.-F., Conway A., Kan J.L.C., Gibson N.W., Willson J.K.V., Cowell J.K., Brattain M.G.
Colon carcinoma cells harboring PIK3CA mutations display resistance to growth factor deprivation induced apoptosis.
Mol. Cancer Ther. 6:1143-1150(2007)
PubMed=18258742; DOI=10.1073/pnas.0712176105; PMCID=PMC2268141
Emaduddin M., Bicknell D.C., Bodmer W.F., Feller S.M.
Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells.
Proc. Natl. Acad. Sci. U.S.A. 105:2358-2362(2008)
PubMed=18340113; DOI=10.4161/cbt.7.6.5838
Gongora C., Candeil L., Vezzio-Vie N., Copois V., Denis V., Bareil C., Molina F., Fraslon C., Conseiller E., Pau B., Martineau P., Del Rio M.
Altered expression of cell proliferation-related and interferon-stimulated genes in colon cancer cells resistant to SN38.
Cancer Biol. Ther. 7:822-832(2008)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921; PMCID=PMC4020356
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=19927377; DOI=10.1002/gcc.20730; PMCID=PMC2818350
Knutsen T., Padilla-Nash H.M., Wangsa D., Barenboim-Stapleton L., Camps J., McNeil N.E., Difilippantonio M.J., Ried T.
Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines.
Genes Chromosomes Cancer 49:204-223(2010)
PubMed=20164919; DOI=10.1038/nature08768; PMCID=PMC3145113
Bignell G.R., Greenman C.D., Davies H.R., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192; PMCID=PMC2943514
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780; PMCID=PMC2920028
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=22068913; DOI=10.1073/pnas.1111840108; PMCID=PMC3219108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=21912889; DOI=10.1007/s10637-011-9744-z
Sutherland H.S., Hwang I.Y., Marshall E.S., Lindsay B.S., Denny W.A., Gilchrist C., Joseph W.R., Greenhalgh D., Richardson E., Kestell P., Ding A., Baguley B.C.
Therapeutic reactivation of mutant p53 protein by quinazoline derivatives.
Invest. New Drugs 30:2035-2045(2012)
PubMed=22336246; DOI=10.1016/j.bmc.2012.01.017
Kong D.-X., Yamori T.
JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs.
Bioorg. Med. Chem. 20:1947-1951(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628; PMCID=PMC3276511
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096; PMCID=PMC3285665
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595; PMCID=PMC3526189
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=23272949; DOI=10.1186/1755-8794-5-66; PMCID=PMC3543849
Schlicker A., Beran G., Chresta C.M., McWalter G., Pritchard A., Weston S., Runswick S., Davenport S., Heathcote K., Castro D.A., Orphanides G., French T., Wessels L.F.A.
Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines.
BMC Med. Genomics 5:66.1-66.15(2012)
PubMed=23546019; DOI=10.3892/ijo.2013.1868
Petitprez A., Poindessous V., Ouaret D., Regairaz M., Bastian G., Guerin E., Escargueil A.E., Larsen A.K.
Acquired irinotecan resistance is accompanied by stable modifications of cell cycle dynamics independent of MSI status.
Int. J. Oncol. 42:1644-1653(2013)
PubMed=23631600; DOI=10.1021/pr400260h
Loftus N.J., Lai L., Wilkinson R.W., Odedra R., Wilson I.D., Barnes A.J.
Global metabolite profiling of human colorectal cancer xenografts in mice using HPLC-MS/MS.
J. Proteome Res. 12:2980-2986(2013)
PubMed=23649806; DOI=10.1083/jcb.201210031; PMCID=PMC3653305
Kleiblova P., Shaltiel I.A., Benada J., Sevcik J., Pechackova S., Pohlreich P., Voest E.E., Dundr P., Bartek J., Kleibl Z., Medema R.H., Macurek L.
Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint.
J. Cell Biol. 201:511-521(2013)
PubMed=23671654; DOI=10.1371/journal.pone.0063056; PMCID=PMC3646030
Lu Y.-H., Soong T.D., Elemento O.
A novel approach for characterizing microsatellite instability in cancer cells.
PLoS ONE 8:E63056-E63056(2013)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342; PMCID=PMC4893961
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24042735; DOI=10.1038/oncsis.2013.35; PMCID=PMC3816225
Ahmed D., Eide P.W., Eilertsen I.A., Danielsen S.A., Eknaes M., Hektoen M., Lind G.E., Lothe R.A.
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis 2:e71.1-e71.8(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20; PMCID=PMC4178206
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24670534; DOI=10.1371/journal.pone.0092047; PMCID=PMC3966786
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893; PMCID=PMC4355981
Boegel S., Lower M., Bukur T., Sahin U., Castle J.C.
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines.
OncoImmunology 3:e954893.1-e954893.12(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35; PMCID=PMC4432652
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25576301; DOI=10.1074/mcp.M114.042812; PMCID=PMC4349985
Bassani-Sternberg M., Pletscher-Frankild S., Jensen L.J., Mann M.
Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation.
Mol. Cell. Proteomics 14:658-673(2015)
PubMed=25841592; DOI=10.1016/j.jprot.2015.03.019
Piersma S.R., Knol J.C., de Reus I., Labots M., Sampadi B.K., Pham T.V., Ishihama Y., Verheul H.M.W., Jimenez C.R.
Feasibility of label-free phosphoproteomics and application to base-line signaling of colorectal cancer cell lines.
J. Proteomics 127:247-258(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=26169745; DOI=10.1186/s12967-015-0576-z; PMCID=PMC4499939
Halama A., Guerrouahen B.S., Pasquier J., Diboun I., Karoly E.D., Suhre K., Rafii A.
Metabolic signatures differentiate ovarian from colon cancer cell lines.
J. Transl. Med. 13:223.1-223.12(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26719794; DOI=10.1186/s13742-015-0106-1; PMCID=PMC4696294
Teo A.S.M., Verzotto D., Yao F., Nagarajan N., Hillmer A.M.
Single-molecule optical genome mapping of a human HapMap and a colorectal cancer cell line.
GigaScience 4:65.1-65.6(2015)
PubMed=26537799; DOI=10.1074/mcp.M115.051235; PMCID=PMC4762531
Holst S., Deuss A.J.M., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A.M., Deelder A.M., Mesker W.E., Tollenaar R.A.E.M., Rombouts Y., Wuhrer M.
N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression.
Mol. Cell. Proteomics 15:124-140(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52; PMCID=PMC4932877
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4; PMCID=PMC5087121
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=28179481; DOI=10.1158/1535-7163.MCT-16-0578
Tanaka N., Mashima T., Mizutani A., Sato A., Aoyama A., Gong B., Yoshida H., Muramatsu Y., Nakata K., Matsuura M., Katayama R., Nagayama S., Fujita N., Sugimoto Y., Seimiya H.
APC mutations as a potential biomarker for sensitivity to tankyrase inhibitors in colorectal cancer.
Mol. Cancer Ther. 16:752-762(2017)
PubMed=28192450; DOI=10.1371/journal.pone.0171435; PMCID=PMC5305277
Fasterius E., Raso C., Kennedy S.A., Rauch N., Lundin P., Kolch W., Uhlen M., Al-Khalili Szigyarto C.
A novel RNA sequencing data analysis method for cell line authentication.
PLoS ONE 12:E0171435-E0171435(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28601559; DOI=10.1016/j.cels.2017.05.009; PMCID=PMC5493283
Bekker-Jensen D.B., Kelstrup C.D., Batth T.S., Larsen S.C., Haldrup C., Bramsen J.B., Sorensen K.D., Hoyer S., Orntoft T.F., Lindbjerg Andersen C., Nielsen M.L., Olsen J.V.
An optimized shotgun strategy for the rapid generation of comprehensive human proteomes.
Cell Syst. 4:587-599.e4(2017)
PubMed=28683746; DOI=10.1186/s12943-017-0691-y; PMCID=PMC5498998
Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A.
Multi-omics of 34 colorectal cancer cell lines -- a resource for biomedical studies.
Mol. Cancer 16:116.1-116.16(2017)
PubMed=28854368; DOI=10.1016/j.celrep.2017.08.010; PMCID=PMC5583477
Roumeliotis T.I., Williams S.P., Goncalves E., Alsinet C., Del Castillo Velasco-Herrera M., Aben N., Ghavidel F.Z., Michaut M., Schubert M., Price S., Wright J.C., Yu L., Yang M., Dienstmann R., Guinney J.H., Beltrao P., Brazma A., Pardo M., Stegle O., Adams D.J., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Choudhary J.S.
Genomic determinants of protein abundance variation in colorectal cancer cells.
Cell Rep. 20:2201-2214(2017)
PubMed=29101300; DOI=10.15252/msb.20177701; PMCID=PMC5731344
Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R.-S., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A., Heinzlmeir S., Johnstone E., Domingo E., Kerr D.J., Jesinghaus M., Slotta-Huspenina J., Weichert W., Knapp S., Feller S.M., Kuster B.
Pharmacoproteomic characterisation of human colon and rectal cancer.
Mol. Syst. Biol. 13:951-951(2017)
PubMed=29207035; DOI=10.3892/ijo.2017.4206
Olejniczak A., Szarynska M., Kmiec Z.
In vitro characterization of spheres derived from colorectal cancer cell lines.
Int. J. Oncol. 52:599-612(2018)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051; PMCID=PMC6343826
Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D.A., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F.
Differential effector engagement by oncogenic KRAS.
Cell Rep. 22:1889-1902(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023; PMCID=PMC7339254
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=32172478; DOI=10.1007/s12253-020-00805-3
Xu Y.-T., Zhang L., Wang Q.-L., Zheng M.-J.
Comparison of different colorectal cancer with liver metastases models using six colorectal cancer cell lines.
Pathol. Oncol. Res. 26:2177-2183(2020)
PubMed=32927768; DOI=10.3390/cancers12092582; PMCID=PMC7564713
Schulte am Esch J., Windmoller B.A., Hanewinkel J., Storm J., Forster C., Wilkens L., Kruger M., Kaltschmidt B., Kaltschmidt C.
Isolation and characterization of two novel colorectal cancer cell lines, containing a subpopulation with potential stem-like properties: treatment options by MYC/NMYC inhibition.
Cancers (Basel) 12:2582.1-2582.34(2020)"
文献支持
HCT 116人结肠癌传代细胞|送STR图谱
¥850 - 2150