相关产品推荐更多 >
万千商家帮你免费找货
0 人在求购买到急需产品
- 详细信息
- 文献和实验
- 技术资料
- 英文名:
RD-ES
- 库存:
100
- 供应商:
中乔新舟
- 品系:
细胞系
- 运输方式:
常温
- 年限:
液氮长期
- 生长状态:
贴壁和悬浮
- 规格:
T25
|
产品名称 |
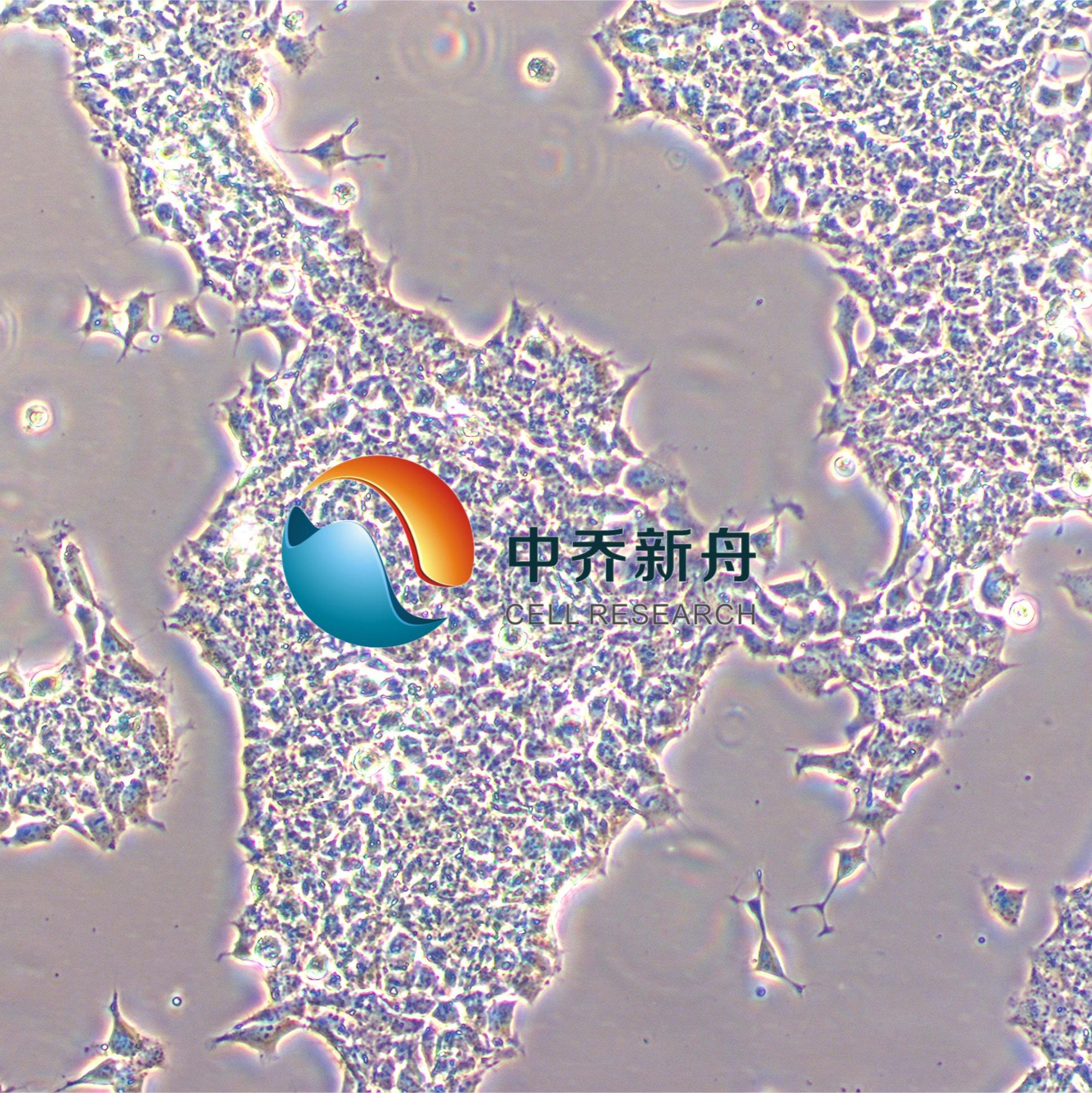
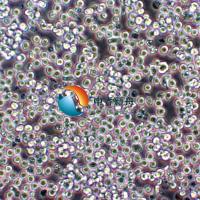
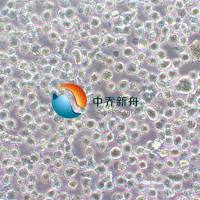
RD-ES 人尤文氏肉瘤 |
|
货号 |
ZQ1018 |
|
产品介绍 |
RD-ES 是一种上皮细胞系,该细胞系由 G. Marshall 和 M. Kirchen 分离自患有尤文氏肉瘤的 19 岁男性 White 的肱骨原发性骨性尤文氏肉瘤中培养而成。从超微结构来看,这些细胞表现出原始细胞连接,具有糖原池,直径为 20 至 25 微米。这些细胞以 5 至 10 个细胞的小簇的形式松散地生长成单层。 这种细胞系在癌症研究中具有重要的应用价值,尤其是在尤文氏肉瘤的研究领域。尤文氏肉瘤(Ewing's sarcoma)是一种罕见的恶性肿瘤,通常发生在儿童和青少年中,表现为未分化的原发性骨肿瘤,偶尔也会发生在软组织中,即骨外尤文肉瘤(extraosseous Ewing sarcoma, EES)。该细胞系为尤文氏肉瘤的研究提供了一个有价值的工具,有助于更好地理解这种疾病并开发新的治疗方法。 |
|
种属 |
人 |
|
性别/年龄 |
男/19 |
|
组织 |
骨 |
|
疾病 |
尤文氏肉瘤 |
|
细胞类型 |
肿瘤细胞 |
|
形态学 |
上皮 |
|
生长方式 |
混合:贴壁和悬浮 |
|
倍增时间 |
~60 hours (DSMZ=ACC-260) |
|
培养基和添加剂 |
RPMI-1640(品牌:中乔新舟 货号:ZQ-200)+10%胎牛血清(中乔新舟 货号:ZQ500-A)+1%P/S(中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
ZM1018 |
|
生物安全等级 |
BSL-1 |
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
STR位点信息 |
Amelogenin: X,Y CSF1PO :11 D2S1338: 19,20 D3S1358 :15 D5S818 :11 D7S820: 10 D8S1179: 13 D13S317: 11,12 D16S539 :9,11 D18S51: 14,18 D19S433 :13,14 D21S11: 28 FGA: 21,25 Penta D: 9,12 Penta E :11,13 SE33: 17,18 TH01: 7 TPOX :9,11 vWA :17 |
|
抗原表达/受体表达 |
*** |
|
基因表达 |
*** |
|
保藏机构 |
ATCC; HTB-166 |
|
供应限制 |
仅供科研使用 |
上海中乔新舟生物科技有限公司成立于2011年,历经十多年发展,主要专注于细胞生物学产品的研究和开发,专注于为药企、各类科研机构及CRO企业提供符合标准规范的细胞培养服务、细胞培养基、细胞检测试剂盒、细胞培养试剂,胎牛血清和细胞生物学技术服务等。
公司一直致力于为高等院校、研究机构、医院、CRO及CDMO企业提供细胞培养完整解决方案,这些产品旨在满足细胞培养的多样需求,确保实验和研究的有效进行。引用中乔新舟(ZQXZBIO)产品和服务的文献超数千篇。

产品服务
细胞资源:原代细胞、细胞株、干细胞、示踪细胞、耐药株细胞、永生化细胞等基因工程细胞。
试剂产品:胎牛血清、完全培养基(适用于原代细胞及细胞株)、无血清培养基、基础培养基、细胞转染试剂、重组因子、胰酶和双抗等等细胞培养所有实验相关产品。
技术服务:稳转株构建、原代细胞分离、特殊培养基定制服务、细胞检测等。

目前产品已经畅销国内30多个省市,与客户建立长期的合作伙伴关系,共同实现成功。全体员工将不懈努力,继续为科研人员提供优良的产品和服务,致力成为全球细胞培养领域的参与者。
企业愿景
致力于成为国内细胞培养基产业的佼佼者,生物医药领域上游原材料的优良提供商。
企业使命
成长为专业细胞系及原代细胞培养供应商、专业细胞培养基及培养试剂生产商。
企业荣誉


风险提示:丁香通仅作为第三方平台,为商家信息发布提供平台空间。用户咨询产品时请注意保护个人信息及财产安全,合理判断,谨慎选购商品,商家和用户对交易行为负责。对于医疗器械类产品,请先查证核实企业经营资质和医疗器械产品注册证情况。
 文献和实验
文献和实验
PubMed=8221663
Komuro H., Hayashi Y., Kawamura M., Hayashi K., Kaneko Y., Kamoshita S., Hanada R., Yamamoto K., Hongo T., Yamada M., Tsuchida Y.
Mutations of the p53 gene are involved in Ewing's sarcomas but not in neuroblastomas.
Cancer Res. 53:5284-5288(1993)
PubMed=8378080
Kovar H., Auinger A., Jug G., Aryee D.N.T., Zoubek A., Salzer-Kuntschik M., Gadner H.
Narrow spectrum of infrequent p53 mutations and absence of MDM2 amplification in Ewing tumours.
Oncogene 8:2683-2690(1993)
PubMed=8040301; DOI=10.1172/JCI117360
Giovannini M., Biegel J.A., Serra M., Wang J.-Y., Wei Y.-L.H., Nycum L., Emanuel B.S., Evans G.A.
EWS-erg and EWS-Fli1 fusion transcripts in Ewing's sarcoma and primitive neuroectodermal tumors with variant translocations.
J. Clin. Invest. 94:489-496(1994)
PubMed=9738976; DOI=10.1111/j.1349-7006.1998.tb03274.x
Urano F., Umezawa A., Yabe H., Hong W., Yoshida K., Fujinaga K., Hata J.-i.
Molecular analysis of Ewing's sarcoma: another fusion gene, EWS-E1AF, available for diagnosis.
Jpn. J. Cancer Res. 89:703-711(1998)
PubMed=11423975; DOI=10.1038/sj.onc.1204437
Dauphinot L., De Oliveira C., Melot T., Sevenet N., Thomas V., Weissman B.E., Delattre O.
Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression.
Oncogene 20:3258-3265(2001)
PubMed=18082704; DOI=10.1016/j.jpedsurg.2007.08.026
Komuro H., Saihara R., Shinya M., Takita J., Kaneko S., Kaneko M., Hayashi Y.
Identification of side population cells (stem-like cell population) in pediatric solid tumor cell lines.
J. Pediatr. Surg. 42:2040-2045(2007)
PubMed=18160777; DOI=10.1159/000109614
Savola S., Nardi F., Scotlandi K., Picci P., Knuutila S.
Microdeletions in 9p21.3 induce false negative results in CDKN2A FISH analysis of Ewing sarcoma.
Cytogenet. Genome Res. 119:21-26(2007)
PubMed=19787792; DOI=10.1002/gcc.20717
Ottaviano L., Schaefer K.-L., Gajewski M., Huckenbeck W., Baldus S.E., Rogel U., Mackintosh C., de Alava E., Myklebost O., Kresse S.H., Meza-Zepeda L.A., Serra M., Cleton-Jansen A.-M., Hogendoorn P.C.W., Buerger H., Aigner T., Gabbert H.E., Poremba C.
Molecular characterization of commonly used cell lines for bone tumor research: a trans-European EuroBoNet effort.
Genes Chromosomes Cancer 49:40-51(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=21822310; DOI=10.1038/onc.2011.317
Mackintosh C., Ordonez J.L., Garcia-Dominguez D.J., Sevillano V., Llombart-Bosch A., Szuhai K., Scotlandi K., Alberghini M., Sciot R., Sinnaeve F., Hogendoorn P.C.W., Picci P., Knuutila S., Dirksen U., Debiec-Rychter M., Schaefer K.-L., de Alava E.
1q gain and CDT2 overexpression underlie an aggressive and highly proliferative form of Ewing sarcoma.
Oncogene 31:1287-1298(2012)
PubMed=22142829; DOI=10.1158/1078-0432.CCR-11-2056
Shukla N., Ameur N., Yilmaz I., Nafa K., Lau C.-Y., Marchetti A., Borsu L., Barr F.G., Ladanyi M.
Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways.
Clin. Cancer Res. 18:748-757(2012)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=25010205; DOI=10.1371/journal.pgen.1004475
Brohl A.S., Solomon D.A., Chang W., Wang J.-J., Song Y., Sindiri S., Patidar R., Hurd L., Chen L., Shern J.F., Liao H.-L., Wen X.-Y., Gerard J., Kim J.-S., Lopez Guerrero J.A., Machado I., Wai D.H., Picci P., Triche T.J., Horvai A.E., Miettinen M.M., Wei J.S., Catchpoole D., Llombart-Bosch A., Waldman T., Khan J.
The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation.
PLoS Genet. 10:E1004475-E1004475(2014)
PubMed=25223734; DOI=10.1158/2159-8290.CD-14-0622
Tirode F., Surdez D., Ma X.-T., Parker M., Le Deley M.-C., Bahrami A., Zhang Z.-J., Lapouble E., Grossetete-Lalami S., Rusch M., Reynaud S., Rio-Frio T., Hedlund E., Wu G., Chen X., Pierron G., Oberlin O., Zaidi S., Lemmon G., Gupta P., Vadodaria B., Easton J., Gut M., Ding L., Mardis E.R., Wilson R.K., Shurtleff S., Laurence V., Michon J., Marec-Berard P., Gut I.G., Downing J.R., Dyer M.A., Zhang J.-H., Delattre O.
Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations.
Cancer Discov. 4:1342-1353(2014)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26351324; DOI=10.1158/1535-7163.MCT-15-0074
Teicher B.A., Polley E.C., Kunkel M., Evans D., Silvers T.E., Delosh R.M., Laudeman J., Ogle C., Reinhart R., Selby M., Connelly J., Harris E., Monks A., Morris J.
Sarcoma cell line screen of oncology drugs and investigational agents identifies patterns associated with gene and microRNA expression.
Mol. Cancer Ther. 14:2452-2462(2015)
PubMed=26428435; DOI=10.1016/j.ejca.2015.08.020
Sand L.G.L., Scotlandi K., Berghuis D., Snaar-Jagalska B.E., Picci P., Schmidt T., Szuhai K., Hogendoorn P.C.W.
CXCL14, CXCR7 expression and CXCR4 splice variant ratio associate with survival and metastases in Ewing sarcoma patients.
Eur. J. Cancer 51:2624-2633(2015)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=29464090; DOI=10.18632/oncotarget.23815
Spurny C., Kailayangiri S., Altvater B., Jamitzky S., Hartmann W., Wardelmann E., Ranft A., Dirksen U., Amler S., Hardes J., Fluegge M., Meltzer J., Farwick N., Greune L., Rossig C.
T cell infiltration into Ewing sarcomas is associated with local expression of immune-inhibitory HLA-G.
Oncotarget 9:6536-6549(2018)
PubMed=30879952; DOI=10.1016/j.ymthe.2019.02.014
Kailayangiri S., Altvater B., Lesch S., Balbach S.T., Gottlich C., Kuhnemundt J., Mikesch J.-H., Schelhaas S., Jamitzky S., Meltzer J., Farwick N., Greune L., Fluegge M., Kerl K., Lode H.N., Siebert N., Muller I., Walles H., Hartmann W., Rossig C.
EZH2 inhibition in Ewing sarcoma upregulates GD2 expression for targeting with gene-modified T cells.
Mol. Ther. 27:933-946(2019)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
DOI=10.5282/edoc.27750
Orth M.F.
Systematic multi-omics profiling of Ewing sarcoma cell lines.
Thesis PhD (2021), Ludwig Maximilians University of Munich, Germany
相关专题 【原理】 限制性显示 PCR 技术 (Restriction Display PCR,RD-PCR) 技术的思路是在获得反转录的 cDNA 之后 , 进行 Sau 3AI 酶切? Sau 3AI 识别位点是四个碱基 , 理论上计算平均每 256 个碱基有一个识别位点 , 然后在酶切片段的两端加上通用接头?接头由 15bp 和 21 bp 长的两条互补寡核苷酸 单链逐步退火形成 , 接头的一端
【分享】2007新书 Primer of Genetic Analysis: A Problems Approach, 3rd ed
Primer of Genetic Analysis: A Problems Approach, 3rd ed Product Details »Book Publisher: Cambridge University Press (01 October, 2007) »ISBN: 052160365X »Book author: Jr, James N. Thompson, Jenna J. Hellack, Gerald Braver, David S. Durica
尤文氏肉瘤又称为未分化网状细胞瘤。原发于骨髓内的原始细胞,是常见的骨的恶性肿瘤。常见于10—15岁少年,早期及可发生肺转移,预后差,但放射治疗敏感。 临床表现 1.主要症状是局部进行性疼痛,逐渐加重。局部肿胀,皮温增高,可扪及肿块,压痛广泛。 2.病程进展快,有体温增高,血白细胞增多,血沉增快,纳差,消瘦等全身症状。 3.可能有肺转移或其他部位骨转移源。 4.X线检查:骨干或干骺端骨质破坏较广泛,呈虫蚀样,可有葱皮样骨膜反应,可有软组织肿块阴影
 技术资料
技术资料暂无技术资料 索取技术资料